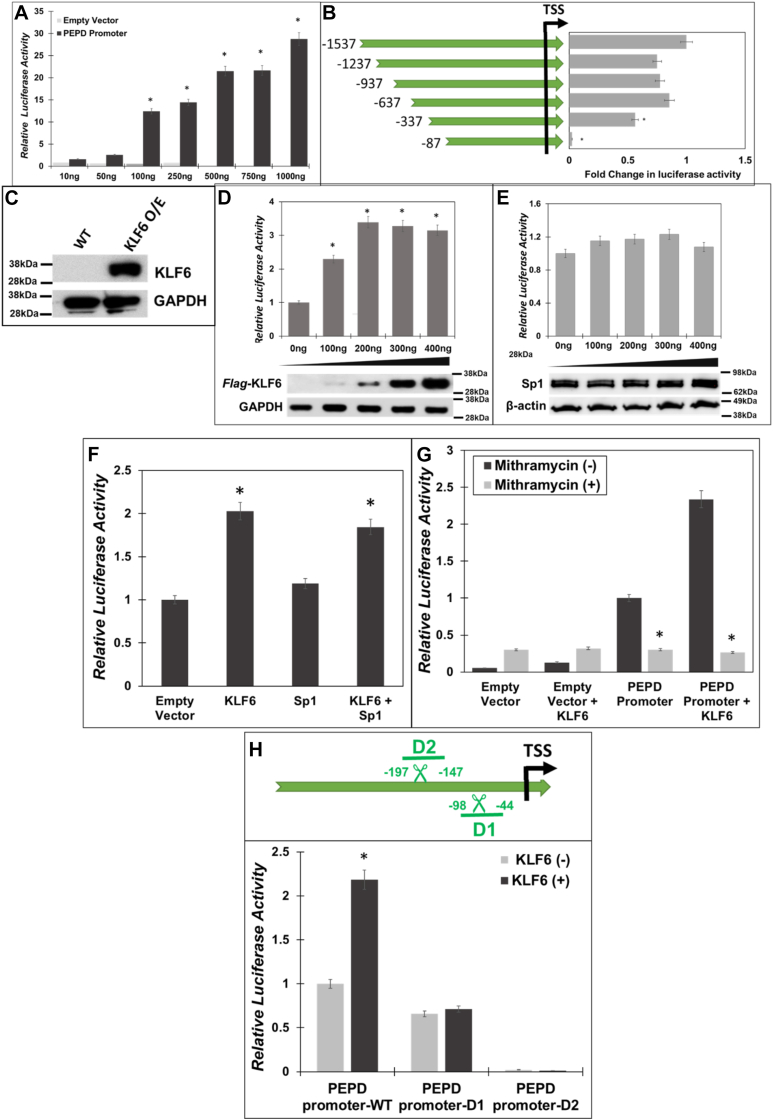
Figure 2.
Sp1 and KLF6 drive PEPD promoter activity.A, the human PEPD promoter region (−1537 bp to +50 bp relative to the TSS) was cloned into the pGL3-basic luciferase reporter (PEPD-Luc). HEK293T cells were co-transfected with increasing amounts of PEPD-Luc or PEPD-null and Renilla Luciferase (pRL-null). Post transfection (24 h), the firefly luciferase activity was measured and normalized to Renilla luciferase activity as an internal control. B, mapping of minimal PEPD promoter: Schematic representation of sequential deletions of PEPD promoter from the 5′-end. Several truncated PEPD-Luc constructs were generated and transfected along with pRLnull into HEK293T cells. Luciferase activity was normalized relative to untruncated PEPD-Luc. C, expression of KLF6 in HEK293T cells without and with the overexpression construct. D–F, HEK293T cells were co-transfected with PEPD-Luc and increasing amounts of either human Flag-KLF6 or Sp1 expression plasmids or both. Then luciferase activity was measured in the cellular lysates and were plotted relative to the control cells. G, HEK293T cells were pretreated with 1 μM of Mith A or vehicle (DMSO) for 1 h, following which the cells were co-transfected with PEPD-Luc with and without human Flag-KLF6. 24 h post transfection the luciferase activity was measured in the cellular lysates and were plotted relative to controls. H, schematic of the two deletion constructs that contained the overlapping KLF6/Sp1 binding sites (upper panel). deletions between – 98 bp and −44 bp (delta 1-D1) and −197 bp to −147 bp (delta 2-D2) relative to TSS of PEPD promoter construct were introduced. These constructs were co-transfected in HEK293T cells with and without human Flag-KLF6. 24 h post transfection the luciferase activity was measured in the cellular lysates and the normalized fold change in luciferase activity was determined. Data are mean values of three independent experiments with error bars representing SEM. ∗ represents a p value of < 0.05 for the statistical comparison of (A and B) empty vector vs PEPD-Luc, (D) empty vector vs PEPD-Luc and KLF6 expression construct, (E) empty vector vs PEPD-Luc and Sp1 expression construct (F) empty vector vs KLF6 or KLF6/Sp1 expression constructs, (G) Untreated vs Mith A treated, and (H) control vs KLF6 expression construct.