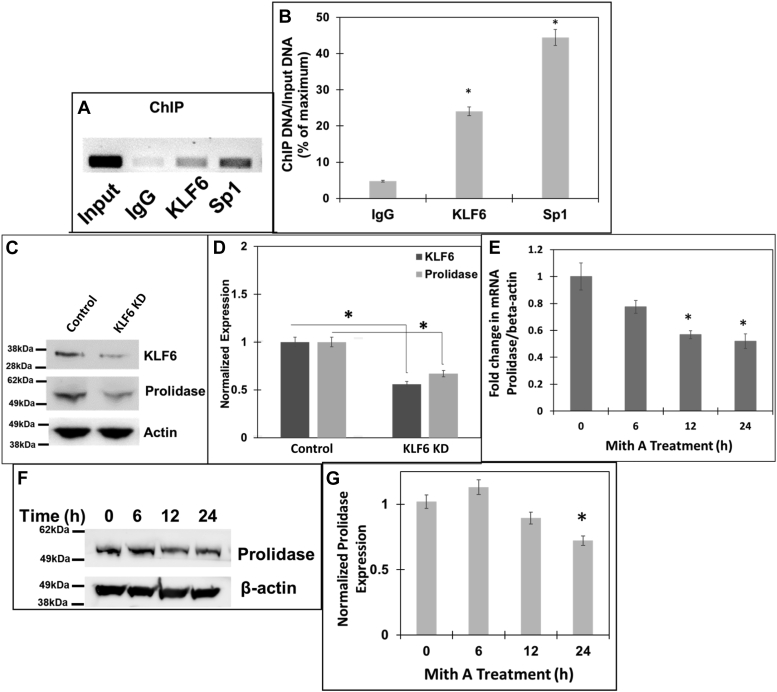
Figure 3.
Role of Sp1 and KLF6 in driving expression of prolidase.A and B, chromatin immunoprecipitation (ChIP) assay of PEPD promoter showing binding of Sp1 and KLF6. Soluble chromatin from (NIH3T3 cells was subjected to ChIP using either KLF6 or Sp1 antibodies or IgG control antibody. Immuno-precipitates were analyzed by PCR with specific primers for the PEPD promoter region encompassing the overlapping Sp1/KLF6 binding sites. A, the PCR products were detected by agarose gel electrophoresis. The input represented the DNA in cell extracts prior to immunoprecipitation. B, densitometric analysis of the immunoprecipitated DNA pulled down with the respective antibodies and are normalized to input from three independent experiments. C and D, NIH3T3 cells were transfected with either non-targeting or KLF6 targeting ASO and then 24 h post transfection, cellular lysates were prepared for Western blot. C, Representative blot (n = 3) showing expression of KLF6, prolidase and β-actin, in KLF6 knocked-down and control cells. D, densitometric analysis of the immunoblot of KLF6 and prolidase that are normalized to β-actin levels. E–G, NIH3T3 cells were treated with 1 μM Mith A or vehicle (DMSO) in a time-dependent manner. E, prolidase mRNA expression in these cells was measured by qPCR and normalized to β-actin mRNA. Prolidase mRNA expression is represented as fold change in Mith A-treated vs untreated cells based on ΔΔCt values. F, equal protein amounts of cell lysates were subjected to immunoblot to measure prolidase protein expression. Representative immunoblot (F) and densitometry analyses (G) of prolidase expression normalized to β-actin protein levels. Data are mean values of three independent experiments with error bars representing SEM. ∗ represents a p value of < 0.05 for the statistical comparison of (B) IgG control vs KLF6 or Sp1, (D) Non-target siRNA control vs KLF6 siRNA, (E and G) untreated vs Mith A treated samples.