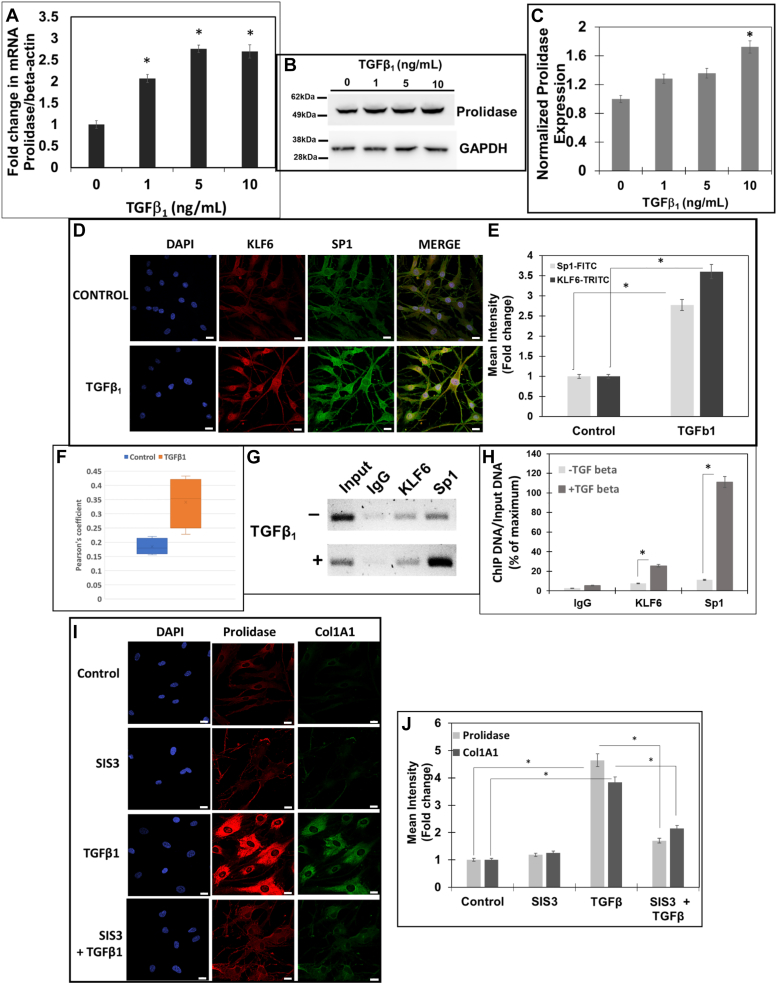
Figure 8.
TGF-β1 induced KLF6/SP1/prolidase axes increase Col1A1 expression in primary human dermal fibroblasts. Serum-starved primary HDFs were treated with increasing concentrations of TGF-β1 (0–10 ng/ml). A, prolidase mRNA levels normalized to GAPDH analyzed by qPCR. B and C, Prolidase protein expression was analyzed by Western blot using GAPDH as a loading control. B, a representative blot, and (C) Densitometric analyses of prolidase expression normalized to GAPDH. D and E, confocal microscopy of KLF6 and Sp1 expression. HDFs were treated with TGF-β1 (10 ng/ml), were fixed/permeabilized and stained with DAPI, anti-KLF6 (red) and anti- Sp1 (green). D, representative images showing KLF6 and Sp1 and colocalization (merge). Scale bars: 20 μm. E, calculated mean fluorescence intensity showing expression of both KLF6 and Sp1 following TGF-β1 treatment as compared to control cells. F, Pearson’s coefficient showing the co-localization of KLF6 and Sp1 following TGF-β1 treatment as compared to control cells. G and H, ChIP was performed using anti-KLF6, anti-Sp1 and IgG (control) antibodies. Co-precipitated chromatin fragments were purified and analyzed by PCR using primers (Table 1) against the human PEPD promoter. PCR products were resolved by agarose gel electrophoresis. Results were normalized to total input chromatin. G, representative blot and (H) Densitometry analyses of PCR amplified DNA. (I and J) HDFs were treated with either vehicle (DMSO) or SIS3 (10 μM) 1 h prior to stimulation with TGF-β1 (10 ng/ml) for 6 h. I, representative confocal images of cells stained for prolidase (red) and Col1A1 (green) and the nuclei (blue). Scale bar, 20 μm. J, the graph represents the mean fluorescence intensity of prolidase and Col1A1 in SIS3-treated vs untreated cells in the presence and absence of TGF-β1. Data are mean values of three independent experiments with error bars representing SEM. ∗ p value of < 0.05 for the statistical comparison of (A–C, E–F, and G) untreated vs TGF-β1 treated samples and (E) untreated control vs TGF-β1 treated samples and TGF-β1 treated vs TGF-β1 + SIS3 treated samples.