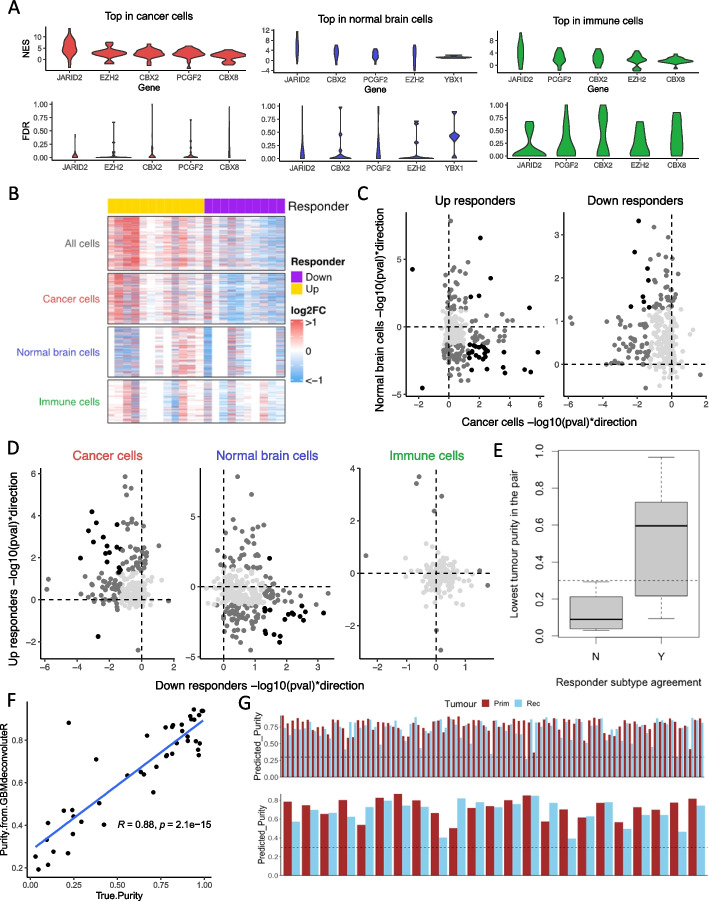
Fig. 3.
A Per-patient normalized enrichment scores (NES, top plot) and false discovery rates (FDR, bottom plot) for the top-scoring promoter-binding factors associated with longitudinal gene expression changes in purified pseudo-bulk samples formed by combining single-cell profiles for: cancer cells (left), normal brain cells (middle), and immune cells (right). B Heatmaps showing the longitudinal fold-change in expression (Log2FC) of LE50 genes (rows) in pseudo-bulk samples consisting of all cells, or purified cell subsets. The responder subtype for each patient (columns), derived from the “all cell” pseudo-bulk, is indicated by the top color bar. C, D LE50 genes plotted according to their direction and significance of dysregulation through treatment in cancer cells (x-axis) and normal brain cells (y-axis) when differential expression analysis is performed separately in Up (left) and Down (right) responders (C); or in Down responders (x-axis) versus Up responders (y-axis) when differential expression analysis is performed separately in pseudo-bulk samples of pure cancer cells (left), normal brain cells (middle), or immune cells (right) (D). Light gray: p > 0.05 in both comparison; dark gray: p < 0.05 in one; black: p < 0.05 in both. E Boxplots of lowest tumor purity (cancer cells as a proportion of all cells) in a longitudinal pair split by to whether that patient’s responder subtype agreed between the cancer cell subset and full tumor pseudo-bulk (Y) or not (N). Dotted line = 30% tumor purity. F True purity of each GBM sample plotted against the purity predicted by applying GBMdeconvoluteR and using the resulting scores in the formula (MES + AC)/(MES + AC + B + DC + Mast + NK + T + Oligodendrocytes). Shaded area: 95% confidence interval. G Predicted tumor purity for Discovery cohort (top) and the Validation cohort (bottom) samples. Dotted line: 30% tumor purity