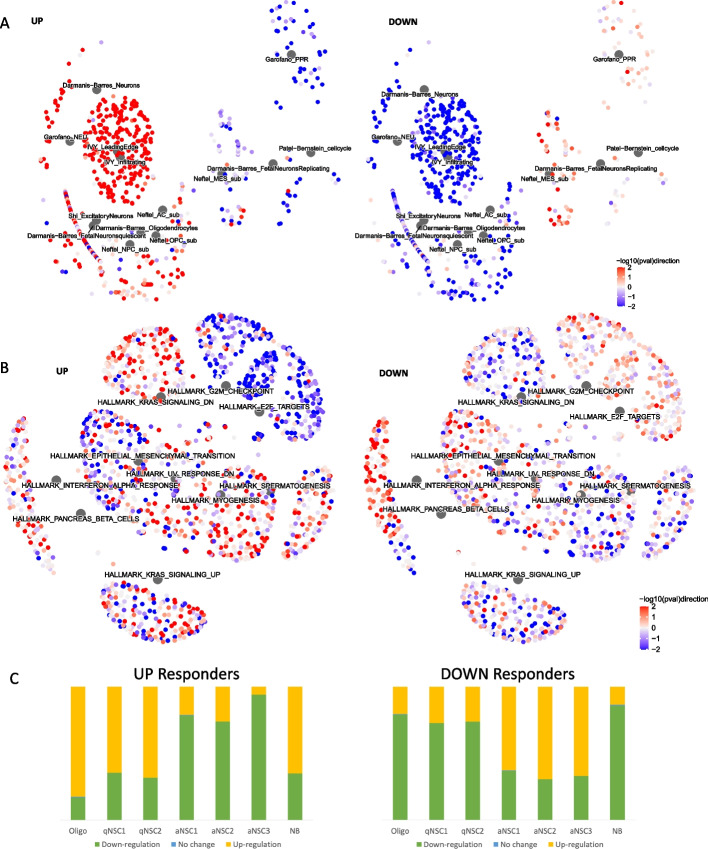
Fig. 4.
A Network plots showing the GBM biology-specific gene sets (described in Additional file 1: Table S24) that are significantly enriched (FDR < 0.05) in either, or both, Up or Down responders through treatment. Large gray hub nodes indicate the gene sets. These have associated, smaller, leaf nodes signifying the genes in that set, colored according to the strength and direction of differential expression through treatment (quantified as -log10p-value multiplied by the direction of fold change: -log10(pval)direction) in Up responders (left image) or Down responders (right image). B As for panel C but visualizing the Hallmark gene sets from MSigDB that were enriched with an FDR < 0.25 in either, or both, responder subtypes. C The proportion of neural stem cell markers of quiescence (qNSC) or active cycling (aNSC), or markers of more differentiated neuroblasts (NB) or oligodendrocytes (Oligo), that were upregulated (yellow), downregulated (green), or stable (blue) in Up (left) and Down (right) responders