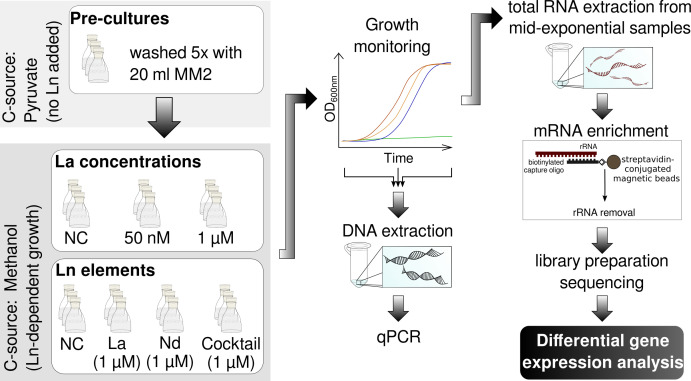
Fig 1.
Overview of the cultivation setup and workflow for the carried out RNAseq experiments. Pre-cultures of Beijerinckiaceae bacterium RH AL1 were grown with pyruvate (0.2%, wt/vol, 18.175 mM) and washed with basal MM2 before being used as inoculum for two sets of incubations. Methanol (0.5%, vol/vol, 123 mM) was used as the carbon source for both sets, one investigating the effect of different (i) La concentrations (50 nM vs 1 µM), and one (ii), the effect of different Ln elements [La vs Nd vs Ln cocktail (Ce, Nd, Dy, Ho, Er, Yb)]. Cultivations were performed in triplicates (n = 3). Medium MM2 supplemented with methanol but without Ln source served as negative control (NC). Samples for DNA extraction and downstream quantitative PCR (qPCR) to determine cell numbers based on lanM gene copies were taken at the beginning and end of the incubation and during mid- to late-exponential phase. Biomass samples for RNA extraction were taken during mid- to late-exponential growth as well. Total RNA from each biological triplicate was enriched for mRNA by means of subtractive hybridization, before being subjected to library preparation, and Illumina sequencing. Pre-processed sequencing data were the starting point for differential gene expression analysis.