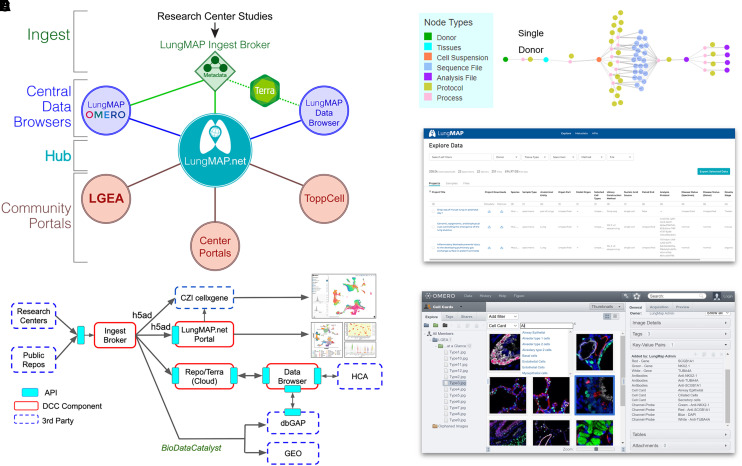
Figure 2.
Advanced omics and image analysis through the LungMAP portal ecosystem. (A) Overview of the LungMAP.net ecosystem, highlighting LungMAP central data browsers for image analysis with the OMERO, dataset browsing through the LungMAP.net graph database or Lung Data Browser Azure database, and connected community portals (Terra, LGEA, ToppCell, ToppGene, and research center portals). (B) Data flow of LungMAP and community data through the centralized LungMAP ingest broker. Standardized metadata is submitted via LungMAP-compatible APIs for deposition in Terra, LungMAP.net, and the LungMAP data browser, as well as controlled-access repositories (dbGAP and BioDataCatalyst) and third-party technologies/portals (ShinyCell, CZI cellxgene, and Azimuth) using LungMAP extended data formats (e.g., h5ad). (C) Standardized metadata graph (on the basis of the HCA metadata schema) for an example biological sample, describing the sample, omics evaluation, protocols, study, and connectivity (process) between entities in the graph. (D) LungMAP data portal to quickly search, download, and reanalyze data (Terra) from LungMAP deposited experiments. (E) LungMAP OMERO image analysis portal to query, explore, combine, reprocess, and export imaging data from diverse imaging modalities. API = application programming interface; CZI = Chan Zuckerberg Initiative; dbGAP = database of Genotypes and Phenotypes; DCC = Data Coordination Center; GEO = Gene Expression Omnibus; HCA = Human Cell Atlas; LGEA = Lung Gene Expression Analysis.