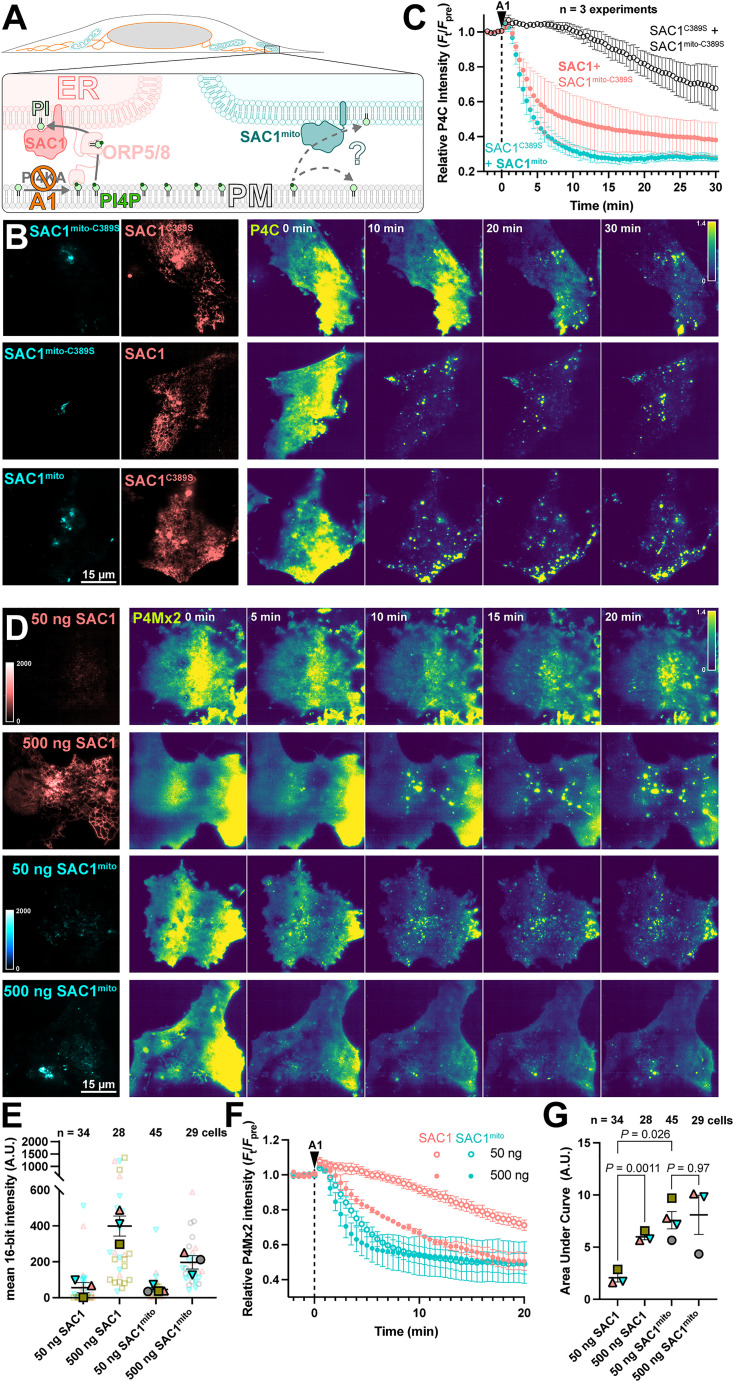
Figure 3.
SAC1mito accelerates PM PI4P turnover. (A) Native SAC1 in the ER is thought to turn over PM PI4P through transfer of the lipid from the PM to the ER; can SAC1mito do the same? (B-C) SAC1mito and over-expressed wild-type SAC1 accelerate PM PI4P turnover. (B) COS-7 cells were transfected with the PI4P biosensor EGFP-P4C-SidC, TagBFP2-SAC1mito, or its inactive C389S mutant, and mCherry-SAC1 or its C389S mutant. Cells were imaged by TIRFM and 30 nM GSK-A1 added to inhibit PM PI4P synthesis at time 0. P4C fluorescence is normalized to the cell's pre-treatement average as indicated. (C) Quantification of the P4C fluorescence data presented in B. Data are grand means ± SE of 3 independent experiments that measured 10-12 cells each. (D-G) SAC1mito is more effective than over-expressed SAC1 at accelerating PM PI4P turnover. (D) COS-7 cells were transfected with the PI4P biosensor mCherry-P4M-SidMx2 and EGFP-SAC1mito or EGFP-SAC1. Images of the EGFP were acquired with identical excitation and detection parameters and are displayed using the same intensity scale. Images are false colored for consistency with B–C. Cells were imaged by TIRFM and 30 nM GSK-A1 added to inhibit PM PI4P synthesis at time 0. (E) Super-plot of the EGFP fluorescence intensity in the TIRFM images; smaller empty symbols show individual cells; larger, filled symbols show the mean of individual experiments. Mean and SE of the 3–4 experiments are shown. (F) Quantification of the P4Mx2 fluorescence intensity from D; data are grand means ± SE of 3–4 experiments. (G) Individual mean area under the curve analysis for individual experiments from F, with mean ± SE indicated. p values are from Tukey's multiple comparisons test after a mixed effects model (P = 0.0089, F [DFn, DFd] = 30.75 [1.584, 3.168], Geisser-Greenhouse's epsilon = 0.5279.