Figure 1.
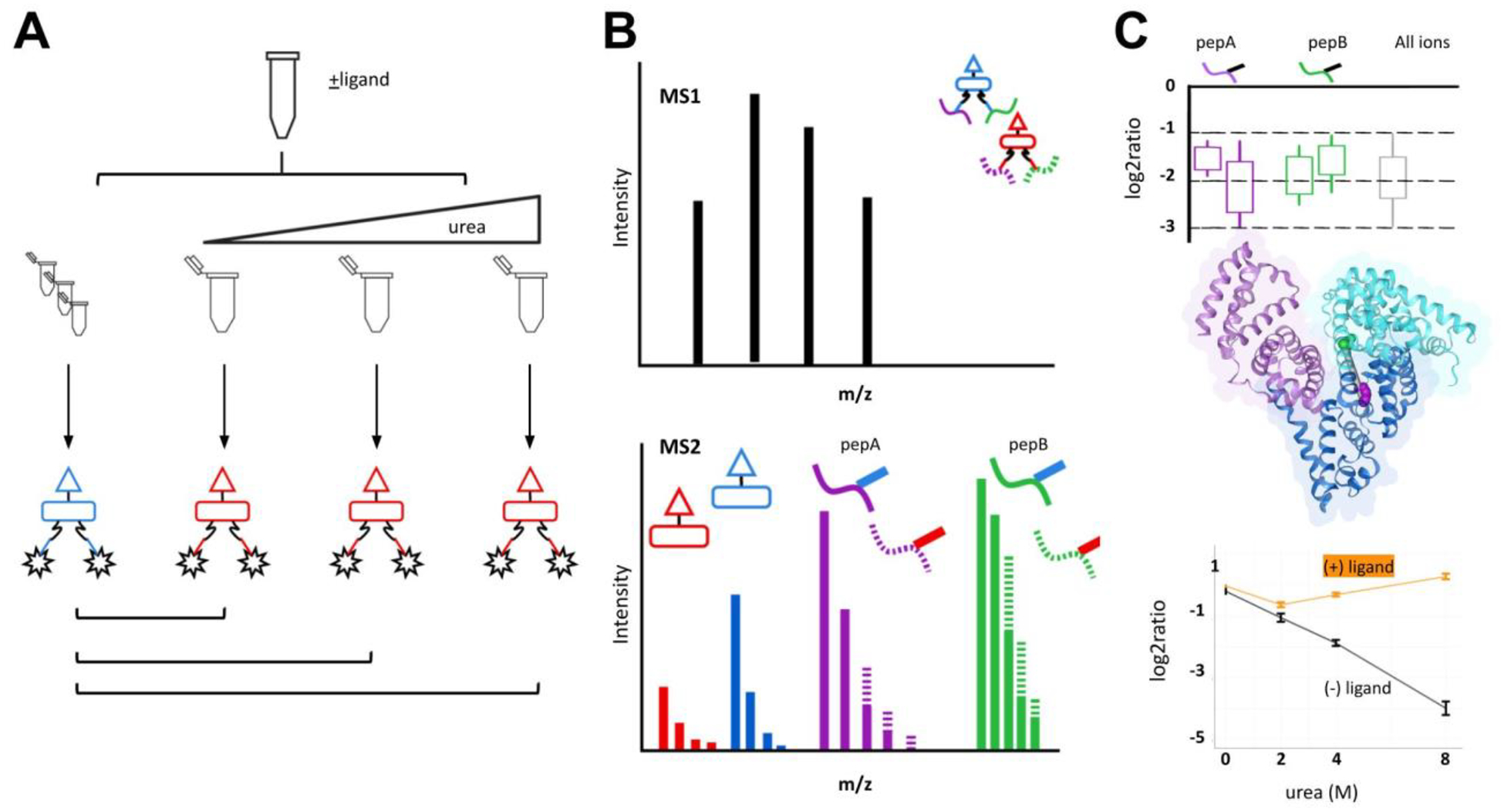
Protein denaturation and quantitative XL-MS workflow.
A) Chemical denaturation of BSA. A BSA stock solution is divided and diluted with increasing urea. Solutions are crosslinked with iqPIR reagent (reporter 808 or 812 m/z). The same crosslinking design was performed for BSA incubated prior with bilirubin. After crosslinking, samples are mixed with a non-denaturing control at a 1:1 ratio. Mixtures are reduced, alkylated, and digested prior to data acquisition. (B) LC-MS. Cross-linked peptide mixtures were prepared for LC-MS/MS using Mango. The data was searched using Comet with crosslink validation using XLinkProphet. (C) Crosslinking data was uploaded to XLinkDB for crosslink peptide pair quantitation viewing and structure mapping. Example peptide K211-K245 (EK211VLTSSAR-LSQK245FPK) was mapped to BSA structure (PDB: 4F5S).
