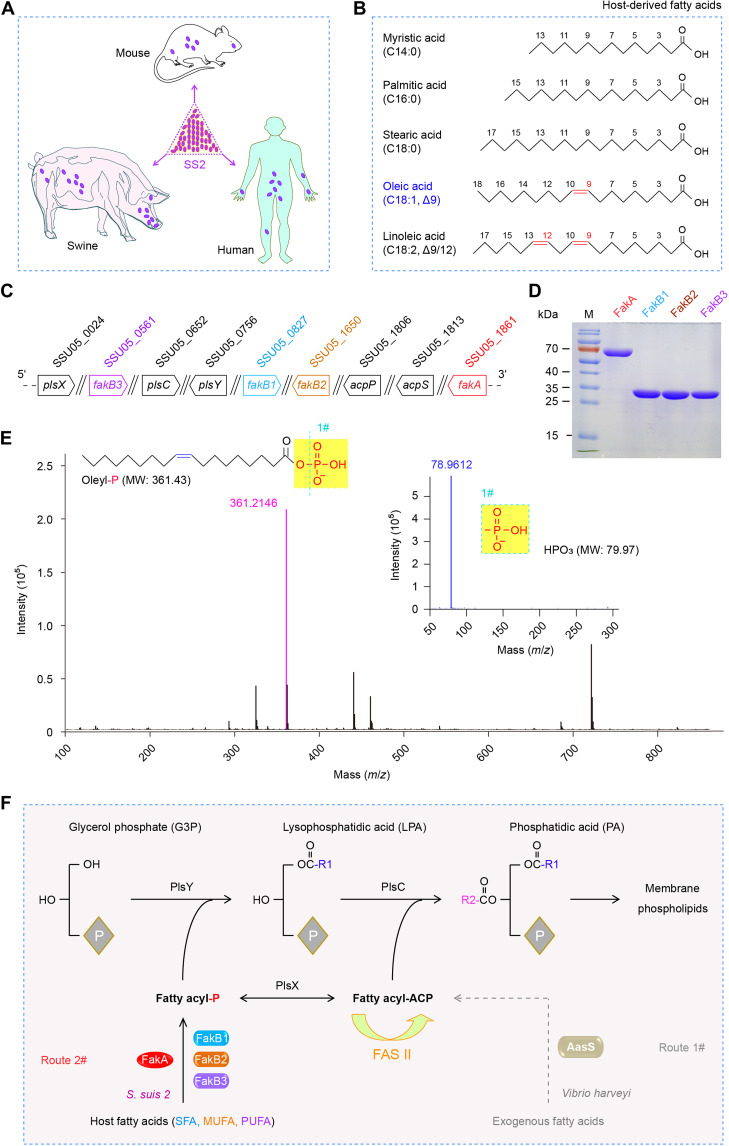
Fig. 1. A scheme for the S. suis FakA-FakB1 (FakB2 and FakB3) system to scavenge host fatty acids.
(A) Cartoon illustration for the zoonotic pathogen S. suis 2 (SS2) and its hosts. Oval purple symbol denotes the bacterium SS2. (B) Chemical structures of five representative host FAs. (C) Genetic environment for the FakA-FakB system and PlsX/Y/C path in SS2. (D) SDS–polyacrylamide gel electrophoresis (PAGE) (12%) profile for four components of the FakA/FakB system from SS2. (E) Use of liquid chromatography–mass spectrometry (LC-MS) to analyze the phosphorylated oleic acid generated by the FakA-FakB2 kinase system. MW, molecular weight. (F) Proposed model of host FA activation by the S. suis FakA-FakB2 (FakB1 and FakB3) system, before the entry into the PlsX/Y/C-based steps of membrane phospholipid formation. Unlike the FakA-FakB2 (FakB1 and FakB3) system of the Gram-positive Streptococcus, the Gram-negative bacterium V. harveyi develops the AasS system to activate host/environmental FAs. Oleyl-P, oleyl-phosphate; acpS, the locus that encodes holo-ACP synthase/phosphopantetheinyl transferase; acpP, acyl carrier protein-encoding gene.