In the title coumarin derivative, the dihedral angle between the 2H-chromen-2-one ring system and the phenyl ring is 89.12 (5)°. In the crystal, the molecules are linked by C—H⋯O hydrogen bonds into [010] double chains.
Keywords: coumarin derivative, Hirshfeld surface, herringbone packing, crystal structure
Abstract
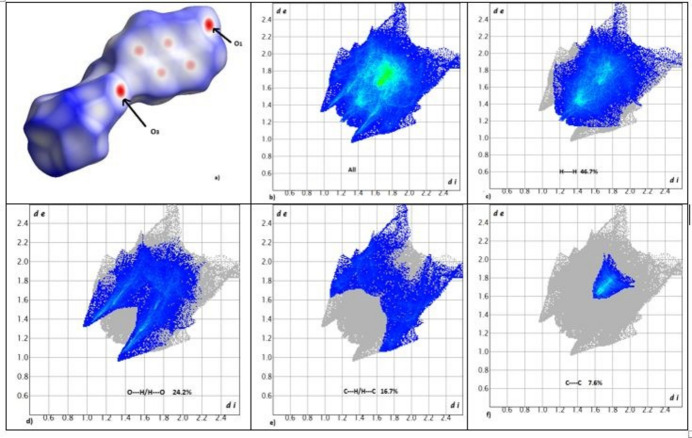
In the title compound, C20H18O4, the dihedral angle between the 2H-chromen-2-one ring system and the phenyl ring is 89.12 (5)°. In the crystal, the molecules are connected through C—H⋯O hydrogen bonds to generate [010] double chains that are reinforced by weak aromatic π–π stacking interactions. The unit-cell packing can be described as a tilted herringbone motif. The H⋯H, H⋯O/O⋯H, H⋯C/C⋯H and C⋯C contacts contribute 46.7, 24.2, 16.7 and 7.6%, respectively, to its Hirshfeld surface.
1. AFRAMED and chemical context
The AFRAMED (Supporting research and training in Africa through remote measurements; Abdel-Aal et al., 2023 ▸) CNRS project was developed by the Chair of the IUCr Africa Initiative (Professor Claude Lecomte) and his team for Crystallography Education in Africa. The project is based on the remote control by an African laboratory of a diffractometer based in France (in fact now at CRM2) to perform X-ray single-crystal diffraction measurements for research and teaching purposes. Selected crystals are sent to the French partner by African researchers who control the data collection remotely and then receive the intensity data by e-mail. The project was launched in August 2022 and is co-financed by the French Centre National de la Recherche Scientifique (CNRS), the United Nations Educational, Scientific and Cultural Organization (UNESCO), and the International Union of Crystallography (IUCr). Two main steps define AFRAMED: first, four weeks training of African Partners (young lecturers with permanent positions) on a single-crystal diffractometer, and in the second step, the African researchers’ laboratories are focal points to assist their colleagues for remote measurements. To date, representatives of Algeria, Cameroon; Congo Brazzaville; Cote d’Ivoire, Egypt, Gabon and Senegal have been trained at the CRM2 laboratory of the Université de Lorraine, France.
This paper presents one of the results of this training: the synthesis, crystal structure and Hirshfeld surface analysis of the title coumarin derivative, I, synthesized by colleagues from Burkina Faso. Such coumaruin derivatives have various biological activities such as anticancer (Lacy et al., 2004 ▸; Kostova, 2005 ▸), anti-inflammatory (Todeschini et al., 1998 ▸), antiviral (Borges et al., 2005 ▸), anti-malarial (Agarwal et al., 2005 ▸), anti-glaucoma (Ziki et al., 2023 ▸) and anticoagulant (Maurer et al., 1998 ▸) properties.
2. Structural commentary
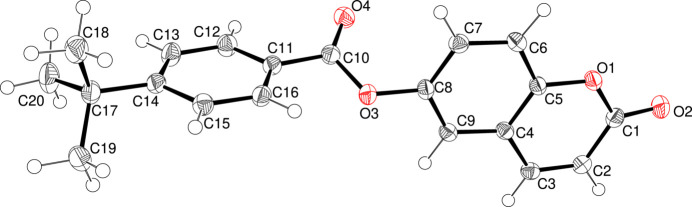
As shown in Fig. 1 ▸, the C1–C9/O1/O2 2H-chromen-2-one ring system of I is almost planar (r.m.s. deviation = 0.044 Å) and the dihedral angle between this ring system and the C11–C16 phenyl group in the 4-tert-butylbenzoate moiety is 89.12 (5)°. This near perpendicular orientation has been observed in other coumarin derivatives with the same motif (Ji et al., 2016 ▸). The dihedral angles between the linking C10/C11/O3/O4 ester group and the pendant C1–C9/O1/O2 and C11–C16 groupings are 64.38 (5) and 25.05 (6)°, respectively, indicating that the major twist in the molecule occurs about the C8—O3 bond. An inspection of the bond lengths shows that there is a slight asymmetry of electronic distribution around the coumarin ring: the difference between the C2=C3 [1.343 (2) Å] and C1—C2 [1.449 (2) Å] separations confirms the double-bond character of the former as indicated in the chemical scheme. Atom C20 of the tert-butyl group lies close to the plane of its attached ring [deviation = 0.226 (2) Å] whereas C18 and C19 are displaced either side of the ring [deviations = −1.465 (1) and 0.964 (1) Å, respectively].
Figure 1.
The molecular structure of I with displacement ellipsoids drawn at the 50% probability level.
3. Supramolecular features
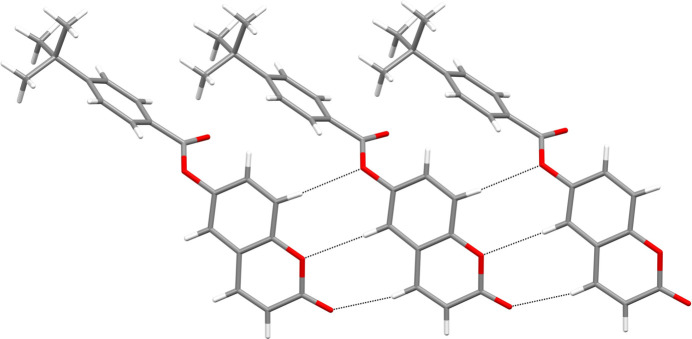
In the crystal, the molecules of I are connected by C—H⋯O hydrogen bonds (Table 1 ▸) to build double chains propagating in the [010] direction: this motif results in two adjacent
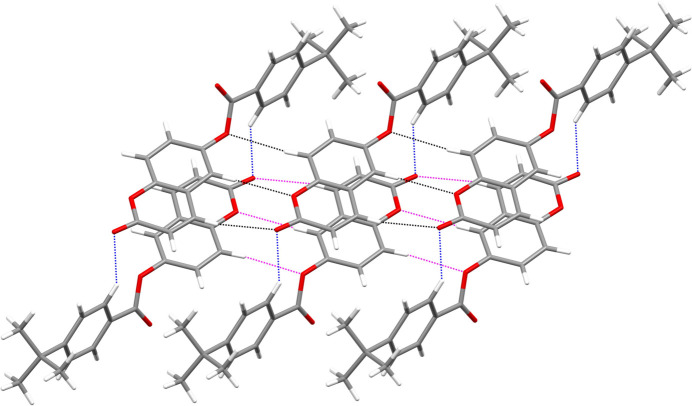
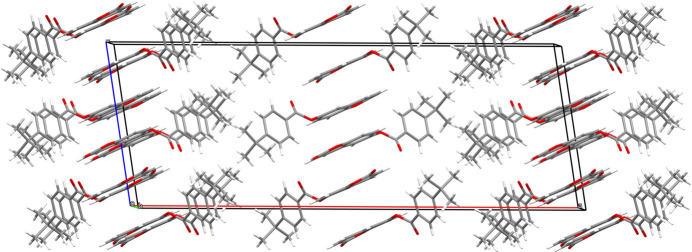
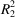 (8)loops between each pair of molecules in one chain formed by the C3, C6 and C9 hydrogen bonds (Fig. 2 ▸). The C16 hydrogen bond provides the linkage to the second chain (Fig. 3 ▸). The pendant 4-tert-butylbenzoate moieties are parallel and shifted by translation along the b axis. Aromatic π–π stacking interactions between centrosymmetric pairs of C4–C9 rings reinforce the cohesion of the double chains [centroid–centroid separation = 3.6301 (8), slippage = 1.579 Å]. The unit-cell packing of I can be described as a tilted herringbone motif (Fig. 4 ▸), as also observed in the crystal structure of 1-(1,2-dihydrophthalazin-1-ylidene)-2-[1-(thiophen-2-yl)ethylidene]hydrazine (Majoumo-Mbe et al., 2019 ▸).
(8)loops between each pair of molecules in one chain formed by the C3, C6 and C9 hydrogen bonds (Fig. 2 ▸). The C16 hydrogen bond provides the linkage to the second chain (Fig. 3 ▸). The pendant 4-tert-butylbenzoate moieties are parallel and shifted by translation along the b axis. Aromatic π–π stacking interactions between centrosymmetric pairs of C4–C9 rings reinforce the cohesion of the double chains [centroid–centroid separation = 3.6301 (8), slippage = 1.579 Å]. The unit-cell packing of I can be described as a tilted herringbone motif (Fig. 4 ▸), as also observed in the crystal structure of 1-(1,2-dihydrophthalazin-1-ylidene)-2-[1-(thiophen-2-yl)ethylidene]hydrazine (Majoumo-Mbe et al., 2019 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C9—H9⋯O1i | 0.987 (13) | 2.584 (13) | 3.5682 (14) | 174.8 (10) |
| C6—H6⋯O3ii | 0.985 (14) | 2.603 (15) | 3.5835 (14) | 173.9 (12) |
| C9—H9⋯O1i | 0.987 (13) | 2.584 (13) | 3.5682 (14) | 174.8 (10) |
| C16—H16⋯O2iii | 0.968 (14) | 2.416 (15) | 3.2628 (16) | 146.0 (11) |
Symmetry codes: (i)
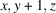 ; (ii)
; (ii)
 ; (iii)
; (iii)
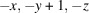 .
.
Figure 2.
Fragment of a [010] chain in the structure of I showing the hydrogen bonds involving C3, C6 and C9 as black dashed lines.
Figure 3.
Partial packing diagram for I showing [010] double chains arising from C—H⋯O hydrogen bonds (black dashed lines in one chain, magenta dashed lines in the other and the C16 cross-linking bonds in blue).
Figure 4.
The unit-cell packing for I viewed down [010].
4. Database survey
A search of the Cambridge Structural Database (CSD, version 5.43; update 3, September 2022; Groom et al., 2016 ▸) for structures having a coumarin motif similar to that of I returned five hits for to the following molecules: 4-methyl-2-oxo-2H-1-benzopyran-6-yl pyridine-2-carboxylate (CSD refcode ATOROT; Ji et al., 2016 ▸), 4-methyl-2-oxo-2H-1-benzopyran-6-yl pyridine-3-carboxylate (ATORUZ; Ji et al., 2016 ▸), 4-methyl-2-oxo-2H-1-benzopyran-6-yl pyridine-4-carboxylate (ATOSAG; Ji et al., 2016 ▸), 6-acetoxycoumarin (GASXON; Murthy et al., 1988 ▸) and 4-methyl-2-oxo-2H-chromen-6-yl benzoate (YEFSOU; Ji et al., 2017 ▸). ATORUZ only features a C6—H6⋯O3 hydrogen bond because a methyl group is bonded to C9 (according to the numbering scheme of I). This prevents the formation of layers like those found in the packing of I, although similar layers are found in GASXON.
5. Hirshfeld surface and Fingerprint plots
The interactions mentioned above are confirmed by the two-dimensional fingerprint plots of I (Fig. 5 ▸). The greatest contributions are the H⋯H and H⋯O/O⋯H contacts with 46.7 and 24.2%, respectively. The H⋯C/C⋯H and C⋯C contacts contribute 16.7 and 7.6%, respectively. The contributions of the H⋯H interactions in I to Hirshfeld surface are greater than those found in 2-oxo-2H-chromen-3-yl 4-chlorobenzoate (Ziki et al. 2017 ▸); this can be related to the packing of the 2H-1-chromen-6-yl moieties of I. The H⋯O/O⋯H contacts are related to the C—H⋯O1 hydrogen bonds shown in Fig. 2 ▸. Their contact points are shown in red and are labelled on the Hirshfeld surface (see Fig. 5 ▸ a).
Figure 5.
(a) Hirshfeld surface of I mapped over d norm and (b) two-dimensional fingerprint plots of (b) overall and delineated into contributions from different contacts: (c) H—H, (d) H—O/O—H, (e) H—C/C—H and (f) C—C.
6. Synthesis and crystallization
To 30 ml solution of 4-tert-butylbenzoyl chloride (1.2 g; 6.17 mmol) in dry tetrahydrofuran, were added dry triethylamine (2.6 ml; 3.1 mmol) and 6-hydroxycoumarin (1.00 g; 6.17 mmol) in small portions over 30 min. The mixture was then refluxed for 4 h and poured into 40 ml of chloroform. The solution was acidified with diluted hydrochloric acid until the pH was 2.5. The organic layer was extracted, washed with water to neutrality, dried over MgSO4 and the solvent removed. The resulting precipitate was suction filtered, washed with petroleum ether and recrystallized from chloroform solution to give colorless prismatic crystals of I in a yield of 84%.
7. Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The H atoms were located in difference maps and their positions and U iso values were freely refined.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C20H18O4 |
| M r | 322.34 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 35.908 (4), 6.8473 (6), 13.2661 (11) |
| β (°) | 98.915 (4) |
| V (Å3) | 3222.3 (5) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.20 × 0.15 × 0.08 |
| Data collection | |
| Diffractometer | Bruker D8 Venture |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.731, 0.895 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 60054, 4940, 3518 |
| R int | 0.061 |
| (sin θ/λ)max (Å−1) | 0.716 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.044, 0.124, 1.11 |
| No. of reflections | 4940 |
| No. of parameters | 289 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.30, −0.27 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989023011052/hb8087sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989023011052/hb8087Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989023011052/hb8087Isup2.cml
CCDC reference: 2301781
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the PMD2 XX-ray diffraction facility (https://crm2.univ-lorraine.fr/lab/fr/services/pmd2x) of the Institut Jean Barriol, Université de Lorraine, for X-ray diffraction measurements and the AFRAMED project. CCDC is also thanked for providing access to the Cambridge Structural Database through the FAIRE program. The authors are very grateful to UNESCO, CNRS and the IUCr for their support to AFRAMED project.
supplementary crystallographic information
Crystal data
| C20H18O4 | F(000) = 1360 |
| Mr = 322.34 | Dx = 1.329 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 4940 reflections |
| a = 35.908 (4) Å | θ = 2.3–30.6° |
| b = 6.8473 (6) Å | µ = 0.09 mm−1 |
| c = 13.2661 (11) Å | T = 100 K |
| β = 98.915 (4)° | Prism, colourless |
| V = 3222.3 (5) Å3 | 0.20 × 0.15 × 0.08 mm |
| Z = 8 |
Data collection
| Bruker D8 Venture diffractometer | 4940 independent reflections |
| Radiation source: fine-focus sealed tube | 3518 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.061 |
| φ and ω scans | θmax = 30.6°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | h = −51→51 |
| Tmin = 0.731, Tmax = 0.895 | k = −9→9 |
| 60054 measured reflections | l = −18→18 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: structure-invariant direct methods |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.124 | All H-atom parameters refined |
| S = 1.11 | w = 1/[σ2(Fo2) + (0.0582P)2 + 0.7544P] where P = (Fo2 + 2Fc2)/3 |
| 4940 reflections | (Δ/σ)max < 0.001 |
| 289 parameters | Δρmax = 0.30 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.01365 (2) | 0.14244 (11) | 0.13861 (6) | 0.02265 (19) | |
| O3 | −0.07925 (2) | 0.77690 (11) | 0.03962 (6) | 0.02359 (19) | |
| C9 | −0.01929 (3) | 0.65096 (16) | 0.10952 (8) | 0.0204 (2) | |
| O2 | 0.06636 (3) | 0.00843 (12) | 0.21536 (7) | 0.0293 (2) | |
| O4 | −0.11921 (2) | 0.70415 (12) | 0.15117 (7) | 0.0276 (2) | |
| C5 | −0.00869 (3) | 0.30453 (15) | 0.11373 (8) | 0.0195 (2) | |
| C2 | 0.06565 (3) | 0.35501 (17) | 0.20298 (9) | 0.0225 (2) | |
| C6 | −0.04569 (3) | 0.26950 (17) | 0.07004 (9) | 0.0219 (2) | |
| C4 | 0.00547 (3) | 0.49326 (15) | 0.13275 (8) | 0.0192 (2) | |
| C1 | 0.05017 (3) | 0.15982 (17) | 0.18837 (9) | 0.0227 (2) | |
| C10 | −0.11073 (3) | 0.80567 (16) | 0.08412 (9) | 0.0222 (2) | |
| C3 | 0.04453 (3) | 0.51431 (17) | 0.17689 (9) | 0.0211 (2) | |
| C11 | −0.13230 (3) | 0.97493 (16) | 0.03591 (9) | 0.0221 (2) | |
| C7 | −0.06982 (3) | 0.42622 (17) | 0.04737 (9) | 0.0221 (2) | |
| C8 | −0.05615 (3) | 0.61479 (16) | 0.06872 (9) | 0.0207 (2) | |
| C16 | −0.12922 (3) | 1.03226 (17) | −0.06341 (9) | 0.0231 (2) | |
| C14 | −0.17523 (3) | 1.28898 (17) | −0.05714 (9) | 0.0235 (2) | |
| C15 | −0.15077 (3) | 1.18512 (17) | −0.10943 (9) | 0.0236 (2) | |
| C12 | −0.15665 (4) | 1.07610 (18) | 0.08908 (10) | 0.0272 (3) | |
| C13 | −0.17742 (4) | 1.23237 (18) | 0.04322 (10) | 0.0280 (3) | |
| C17 | −0.19912 (3) | 1.45327 (17) | −0.11186 (10) | 0.0263 (3) | |
| C19 | −0.17461 (4) | 1.58787 (19) | −0.16687 (12) | 0.0327 (3) | |
| C20 | −0.21829 (5) | 1.5761 (2) | −0.03849 (13) | 0.0402 (4) | |
| C18 | −0.22939 (4) | 1.3594 (2) | −0.19155 (13) | 0.0375 (3) | |
| H9 | −0.0100 (4) | 0.7858 (19) | 0.1225 (9) | 0.018 (3)* | |
| H3 | 0.0540 (4) | 0.644 (2) | 0.1881 (10) | 0.022 (3)* | |
| H2 | 0.0923 (4) | 0.3644 (18) | 0.2369 (9) | 0.019 (3)* | |
| H6 | −0.0534 (4) | 0.132 (2) | 0.0585 (11) | 0.031 (4)* | |
| H16 | −0.1128 (4) | 0.963 (2) | −0.1025 (11) | 0.028 (4)* | |
| H7 | −0.0961 (4) | 0.4084 (19) | 0.0178 (10) | 0.024 (3)* | |
| H15 | −0.1483 (4) | 1.220 (2) | −0.1788 (11) | 0.028 (4)* | |
| H12 | −0.1584 (4) | 1.038 (2) | 0.1596 (12) | 0.033 (4)* | |
| H13 | −0.1935 (4) | 1.302 (2) | 0.0848 (12) | 0.040 (4)* | |
| H19A | −0.1646 (5) | 1.519 (2) | −0.2236 (13) | 0.042 (4)* | |
| H18A | −0.2457 (5) | 1.460 (2) | −0.2292 (12) | 0.042 (4)* | |
| H19B | −0.1528 (5) | 1.642 (2) | −0.1159 (13) | 0.046 (5)* | |
| H20A | −0.2378 (4) | 1.495 (2) | −0.0068 (12) | 0.038 (4)* | |
| H18B | −0.2453 (5) | 1.273 (3) | −0.1618 (14) | 0.054 (5)* | |
| H20B | −0.1987 (4) | 1.634 (2) | 0.0198 (12) | 0.041 (4)* | |
| H20C | −0.2317 (5) | 1.687 (3) | −0.0776 (13) | 0.051 (5)* | |
| H19C | −0.1898 (5) | 1.702 (2) | −0.1991 (12) | 0.045 (4)* | |
| H18C | −0.2170 (5) | 1.281 (3) | −0.2455 (14) | 0.055 (5)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0223 (4) | 0.0180 (4) | 0.0272 (4) | 0.0017 (3) | 0.0023 (3) | 0.0003 (3) |
| O3 | 0.0218 (4) | 0.0213 (4) | 0.0278 (4) | 0.0041 (3) | 0.0043 (3) | 0.0037 (3) |
| C9 | 0.0228 (6) | 0.0186 (5) | 0.0200 (5) | −0.0004 (4) | 0.0039 (4) | 0.0008 (4) |
| O2 | 0.0292 (5) | 0.0237 (4) | 0.0349 (5) | 0.0074 (3) | 0.0046 (4) | 0.0034 (4) |
| O4 | 0.0293 (5) | 0.0255 (4) | 0.0289 (5) | 0.0011 (4) | 0.0069 (4) | 0.0031 (3) |
| C5 | 0.0218 (5) | 0.0173 (5) | 0.0197 (5) | 0.0018 (4) | 0.0044 (4) | 0.0010 (4) |
| C2 | 0.0200 (5) | 0.0251 (6) | 0.0227 (6) | 0.0001 (4) | 0.0042 (4) | −0.0004 (4) |
| C6 | 0.0250 (6) | 0.0184 (5) | 0.0223 (6) | −0.0014 (4) | 0.0035 (4) | −0.0009 (4) |
| C4 | 0.0215 (5) | 0.0182 (5) | 0.0178 (5) | −0.0005 (4) | 0.0033 (4) | 0.0007 (4) |
| C1 | 0.0219 (5) | 0.0245 (6) | 0.0225 (6) | 0.0027 (4) | 0.0058 (4) | 0.0003 (4) |
| C10 | 0.0210 (5) | 0.0211 (5) | 0.0239 (6) | −0.0005 (4) | 0.0017 (4) | −0.0027 (4) |
| C3 | 0.0223 (6) | 0.0215 (5) | 0.0199 (5) | −0.0023 (4) | 0.0045 (4) | −0.0003 (4) |
| C11 | 0.0197 (5) | 0.0206 (5) | 0.0256 (6) | 0.0001 (4) | 0.0023 (4) | −0.0012 (4) |
| C7 | 0.0206 (6) | 0.0226 (5) | 0.0226 (6) | −0.0012 (4) | 0.0021 (4) | 0.0005 (4) |
| C8 | 0.0226 (6) | 0.0188 (5) | 0.0209 (5) | 0.0038 (4) | 0.0042 (4) | 0.0017 (4) |
| C16 | 0.0211 (6) | 0.0227 (5) | 0.0258 (6) | 0.0020 (4) | 0.0043 (4) | −0.0019 (4) |
| C14 | 0.0190 (5) | 0.0207 (5) | 0.0297 (6) | −0.0001 (4) | 0.0010 (4) | −0.0014 (4) |
| C15 | 0.0233 (6) | 0.0240 (6) | 0.0228 (6) | 0.0007 (4) | 0.0015 (4) | −0.0001 (4) |
| C12 | 0.0288 (6) | 0.0282 (6) | 0.0257 (6) | 0.0037 (5) | 0.0072 (5) | 0.0014 (5) |
| C13 | 0.0275 (6) | 0.0277 (6) | 0.0299 (6) | 0.0067 (5) | 0.0081 (5) | −0.0014 (5) |
| C17 | 0.0232 (6) | 0.0229 (6) | 0.0318 (7) | 0.0041 (5) | 0.0008 (5) | −0.0002 (5) |
| C19 | 0.0331 (7) | 0.0242 (6) | 0.0396 (8) | 0.0013 (5) | 0.0017 (6) | 0.0042 (5) |
| C20 | 0.0427 (9) | 0.0341 (7) | 0.0451 (9) | 0.0169 (7) | 0.0104 (7) | 0.0018 (6) |
| C18 | 0.0288 (7) | 0.0307 (7) | 0.0482 (9) | 0.0014 (6) | −0.0094 (6) | 0.0017 (6) |
Geometric parameters (Å, º)
| O1—C1 | 1.3789 (14) | C7—H7 | 0.973 (13) |
| O1—C5 | 1.3791 (13) | C16—C15 | 1.3858 (16) |
| O3—C10 | 1.3680 (14) | C16—H16 | 0.967 (14) |
| O3—C8 | 1.4036 (13) | C14—C15 | 1.3956 (17) |
| C9—C8 | 1.3724 (16) | C14—C13 | 1.4005 (18) |
| C9—C4 | 1.4023 (15) | C14—C17 | 1.5281 (16) |
| C9—H9 | 0.987 (13) | C15—H15 | 0.967 (14) |
| O2—C1 | 1.2150 (14) | C12—C13 | 1.3895 (17) |
| O4—C10 | 1.2047 (14) | C12—H12 | 0.983 (15) |
| C5—C6 | 1.3855 (16) | C13—H13 | 0.983 (16) |
| C5—C4 | 1.3972 (15) | C17—C20 | 1.5286 (19) |
| C2—C3 | 1.3430 (16) | C17—C19 | 1.5347 (19) |
| C2—C1 | 1.4490 (16) | C17—C18 | 1.5348 (18) |
| C2—H2 | 0.993 (13) | C19—H19A | 0.999 (17) |
| C6—C7 | 1.3829 (16) | C19—H19B | 1.021 (17) |
| C6—H6 | 0.985 (14) | C19—H19C | 1.007 (17) |
| C4—C3 | 1.4410 (16) | C20—H20A | 1.034 (16) |
| C10—C11 | 1.4829 (16) | C20—H20B | 1.040 (16) |
| C3—H3 | 0.956 (13) | C20—H20C | 0.999 (18) |
| C11—C12 | 1.3903 (17) | C18—H18A | 0.990 (16) |
| C11—C16 | 1.3955 (17) | C18—H18B | 0.948 (18) |
| C7—C8 | 1.3949 (16) | C18—H18C | 1.047 (18) |
| C1—O1—C5 | 121.32 (9) | C11—C16—H16 | 120.8 (8) |
| C10—O3—C8 | 119.20 (9) | C15—C14—C13 | 117.63 (11) |
| C8—C9—C4 | 119.17 (10) | C15—C14—C17 | 119.29 (11) |
| C8—C9—H9 | 121.0 (7) | C13—C14—C17 | 123.04 (11) |
| C4—C9—H9 | 119.8 (7) | C16—C15—C14 | 121.16 (11) |
| O1—C5—C6 | 116.43 (9) | C16—C15—H15 | 118.5 (8) |
| O1—C5—C4 | 121.28 (10) | C14—C15—H15 | 120.3 (8) |
| C6—C5—C4 | 122.28 (10) | C13—C12—C11 | 119.91 (12) |
| C3—C2—C1 | 121.66 (11) | C13—C12—H12 | 120.9 (8) |
| C3—C2—H2 | 122.0 (7) | C11—C12—H12 | 119.2 (8) |
| C1—C2—H2 | 116.3 (7) | C12—C13—C14 | 121.60 (11) |
| C7—C6—C5 | 118.98 (10) | C12—C13—H13 | 116.8 (9) |
| C7—C6—H6 | 123.8 (8) | C14—C13—H13 | 121.6 (9) |
| C5—C6—H6 | 117.2 (8) | C14—C17—C20 | 112.19 (11) |
| C5—C4—C9 | 118.16 (10) | C14—C17—C19 | 110.30 (10) |
| C5—C4—C3 | 118.03 (10) | C20—C17—C19 | 108.67 (11) |
| C9—C4—C3 | 123.80 (10) | C14—C17—C18 | 107.72 (10) |
| O2—C1—O1 | 116.33 (10) | C20—C17—C18 | 109.17 (12) |
| O2—C1—C2 | 126.25 (11) | C19—C17—C18 | 108.73 (12) |
| O1—C1—C2 | 117.41 (10) | C17—C19—H19A | 111.9 (9) |
| O4—C10—O3 | 123.78 (10) | C17—C19—H19B | 109.9 (9) |
| O4—C10—C11 | 126.47 (11) | H19A—C19—H19B | 110.0 (13) |
| O3—C10—C11 | 109.74 (10) | C17—C19—H19C | 110.8 (9) |
| C2—C3—C4 | 119.92 (10) | H19A—C19—H19C | 106.1 (13) |
| C2—C3—H3 | 122.8 (8) | H19B—C19—H19C | 107.9 (13) |
| C4—C3—H3 | 117.2 (8) | C17—C20—H20A | 111.2 (9) |
| C12—C11—C16 | 119.14 (11) | C17—C20—H20B | 111.4 (9) |
| C12—C11—C10 | 119.88 (11) | H20A—C20—H20B | 109.1 (13) |
| C16—C11—C10 | 120.97 (10) | C17—C20—H20C | 108.3 (10) |
| C6—C7—C8 | 119.02 (11) | H20A—C20—H20C | 108.6 (14) |
| C6—C7—H7 | 121.8 (8) | H20B—C20—H20C | 108.1 (13) |
| C8—C7—H7 | 119.2 (8) | C17—C18—H18A | 110.9 (9) |
| C9—C8—C7 | 122.33 (10) | C17—C18—H18B | 112.4 (11) |
| C9—C8—O3 | 117.29 (10) | H18A—C18—H18B | 107.1 (14) |
| C7—C8—O3 | 120.12 (10) | C17—C18—H18C | 110.8 (10) |
| C15—C16—C11 | 120.53 (11) | H18A—C18—H18C | 107.2 (13) |
| C15—C16—H16 | 118.6 (8) | H18B—C18—H18C | 108.3 (14) |
| C1—O1—C5—C6 | −175.80 (10) | C5—C6—C7—C8 | 0.31 (17) |
| C1—O1—C5—C4 | 3.67 (16) | C4—C9—C8—C7 | −1.38 (17) |
| O1—C5—C6—C7 | 177.35 (10) | C4—C9—C8—O3 | −175.41 (10) |
| C4—C5—C6—C7 | −2.11 (17) | C6—C7—C8—C9 | 1.43 (18) |
| O1—C5—C4—C9 | −177.29 (10) | C6—C7—C8—O3 | 175.30 (10) |
| C6—C5—C4—C9 | 2.15 (17) | C10—O3—C8—C9 | −120.25 (11) |
| O1—C5—C4—C3 | 1.60 (16) | C10—O3—C8—C7 | 65.57 (14) |
| C6—C5—C4—C3 | −178.97 (10) | C12—C11—C16—C15 | −1.48 (18) |
| C8—C9—C4—C5 | −0.39 (16) | C10—C11—C16—C15 | 177.09 (10) |
| C8—C9—C4—C3 | −179.21 (10) | C11—C16—C15—C14 | 1.79 (18) |
| C5—O1—C1—O2 | 173.38 (10) | C13—C14—C15—C16 | −0.42 (17) |
| C5—O1—C1—C2 | −6.99 (15) | C17—C14—C15—C16 | −178.56 (11) |
| C3—C2—C1—O2 | −175.08 (12) | C16—C11—C12—C13 | −0.18 (18) |
| C3—C2—C1—O1 | 5.34 (17) | C10—C11—C12—C13 | −178.76 (11) |
| C8—O3—C10—O4 | 3.51 (16) | C11—C12—C13—C14 | 1.6 (2) |
| C8—O3—C10—C11 | −175.37 (9) | C15—C14—C13—C12 | −1.26 (18) |
| C1—C2—C3—C4 | −0.26 (17) | C17—C14—C13—C12 | 176.81 (12) |
| C5—C4—C3—C2 | −3.21 (16) | C15—C14—C17—C20 | −168.91 (12) |
| C9—C4—C3—C2 | 175.60 (11) | C13—C14—C17—C20 | 13.06 (17) |
| O4—C10—C11—C12 | 24.57 (18) | C15—C14—C17—C19 | −47.60 (15) |
| O3—C10—C11—C12 | −156.59 (11) | C13—C14—C17—C19 | 134.37 (13) |
| O4—C10—C11—C16 | −153.99 (12) | C15—C14—C17—C18 | 70.93 (15) |
| O3—C10—C11—C16 | 24.85 (15) | C13—C14—C17—C18 | −107.11 (14) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C9—H9···O1i | 0.987 (13) | 2.584 (13) | 3.5682 (14) | 174.8 (10) |
| C6—H6···O3ii | 0.985 (14) | 2.603 (15) | 3.5835 (14) | 173.9 (12) |
| C9—H9···O1i | 0.987 (13) | 2.584 (13) | 3.5682 (14) | 174.8 (10) |
| C16—H16···O2iii | 0.968 (14) | 2.416 (15) | 3.2628 (16) | 146.0 (11) |
Symmetry codes: (i) x, y+1, z; (ii) x, y−1, z; (iii) −x, −y+1, −z.
References
- Abdel-Aal, S. K., Kenfack, T. P., Bouraima, A., Djifa, H. A., Emmanuel, W., Bendeif, E.-E. & Lecomte, C. (2023). https://www. iucr. org/news/newsletter/volume-31/number-1/appui-a-la-formation-et-la-recherche-a-travers-les-mesures-a-distance-aframed-a-recent-and-ambitious-project-for-the-development-of-crystallography-in-africa.
- Agarwal, A., Srivastava, K., Puri, S. K. & Chauhan, P. M. S. (2005). Bioorg. Med. Chem. 13, 4645–4650. [DOI] [PubMed]
- 12
- Bruker (2019). APEX3 and SAINT. Bruker AXS Inc. Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Ji, W., Li, L., Eniola-Adefeso, O., Wang, Y., Liu, C. & Feng, C. (2017). J. Mater. Chem. B, 5, 7790–7795. [DOI] [PubMed]
- Ji, W., Liu, G., Li, Z. & Feng, C. (2016). Appl. Mater. Interfaces, 8, 5188–5195. [DOI] [PubMed]
- Kostova, I. (2005). Curr. Med. Chem. Anti-Cancer Agents, 5, 29–46. [DOI] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Lacy, A. & O’Kennedy, R. (2004). Curr. Pharm. Des. 10, 3797–3811. [DOI] [PubMed]
- Majoumo-Mbe, F., Ngwang Nfor, E., Kenfack Tsobnang, P., Nguepmeni Eloundou, V. B., Ngwain Yong, J. & Iris Efeti, I. (2019). Acta Cryst. E75, 251–254. [DOI] [PMC free article] [PubMed]
- Maurer, H. H. & Arlt, J. W. J. (1998). J. Chromatogr. B Biomed. Sci. Appl. 714, 181–195. [DOI] [PubMed]
- Murthy, G. S., Ramamurthy, V. & Venkatesan, K. (1988). Acta Cryst. C44, 307–311.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Todeschini, A. R., de Miranda, A. L. P., da Silva, K. C. M., Parrini, S. C. & Barreiro, E. J. (1998). Eur. J. Med. Chem. 33, 189–199.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Ziki, E., Akonan, L., Kouman, K. C., Dali, D., Megnassan, E., Kakou-Yao, R., Tenon, A. J., Frecer, V. & Miertus, S. J. (2023). Pharm. Res. Int. 35, 10–33.
- Ziki, E., Sosso, S., Mansilla-Koblavi, F., Djandé, A. & Kakou-Yao, R. (2017). Acta Cryst. E73, 45–47. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989023011052/hb8087sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989023011052/hb8087Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989023011052/hb8087Isup2.cml
CCDC reference: 2301781
Additional supporting information: crystallographic information; 3D view; checkCIF report