Abstract
Bone Morphogenic Protein (BMP) signaling plays an essential and highly conserved role in axial patterning in embryos of many externally developing animal species. However, in mammalian embryos, which develop inside the mother, early development includes an additional stage known as preimplantation. During preimplantation, the epiblast lineage is segregated from the extraembryonic lineages that enable implantation and development in utero. Yet, the requirement for BMP signaling in mouse preimplantation is imprecisely defined. We show that, in contrast to prior reports, BMP signaling (as reported by SMAD1/5/9 phosphorylation) is not detectable until implantation, when it is detected in the primitive endoderm – an extraembryonic lineage. Moreover, preimplantation development appears normal following deletion of maternal and zygotic Smad4, an essential effector of BMP signaling. In fact, mice lacking maternal Smad4 are viable. Finally, we uncover a new requirement for zygotic Smad4 in epiblast scaling and cavitation immediately after implantation, via a mechanism involving FGFR/ERK attenuation. Altogether, our results demonstrate no role for BMP4/SMAD4 in the first lineage decisions during mouse development. Rather, multi-pathway signaling among embryonic and extraembryonic cell types drives epiblast morphogenesis post-implantation.
Keywords: pSMAD1/5/9, extraembryonic, maternal and zygotic gene deletion, epiblast, morphogenesis
Summary Statement:
Gene expression, gene deletion, and pathway visualization evidence show that Smad4-dependent signaling is first active after mouse embryo implantation, when it promotes epiblast morphogenesis non-cell autonomously.
Introduction
In animal embryos, including mice, frogs, fish, and flies, the Bone Morphogenic Protein (BMP) signaling pathway oversees critical patterning events early in development. In non-mammalian species, BMP signaling is critical for specification of the dorsal/ventral axis of the early embryo (De Robertis and Sasai, 1996; O’Connor et al., 2006; Zinski et al., 2018). However, the mammalian embryo has an additional developmental task immediately following fertilization: specification of the extraembryonic lineages that will give rise to placenta and yolk sac and enable development within the mother. Published studies support roles for BMP signaling in both extraembryonic lineage specification, prior to implantation, and subsequent axial patterning, which occurs after implantation. However, differences in technical approaches used, as well as challenges intrinsic to mouse, have limited origination of a universally accepted model of the role of BMP signaling in mouse embryos throughout pre- and post-implantation stages.
BMP is one of several related and highly conserved molecular signaling pathways belonging to the Transforming Growth Factor beta (TGFβ) superfamily of cytokines. The molecular mechanisms of TGFβ signaling have been carefully studied (Chang, 2016; Massagué and Sheppard, 2023). BMP proteins, like other members of the TGFβ pathway, are secreted ligands that elicit cellular responses by binding to heterodimeric, transmembrane serine-threonine kinase receptors. The activated receptor complex then phosphorylates members of a family of intracellular effectors known as receptor-associated SMADs (r-SMADs). Phosphorylation of r-SMADs allows their association with a co-factor SMAD and accumulation in the nucleus, where they impact chromatin and transcription (Hill, 2016). In mammals, r-SMAD activity is encoded by several Smad paralogues, with SMAD1, SMAD5, and SMAD9 (also known as SMAD8) primarily transducing BMP signals and SMAD2 and SMAD3 primarily transducing Nodal, Activin, and TGFβ. Notably, the mammalian genome encodes a single co-Smad, SMAD4, which is shared by BMP, Nodal, Activin, and TGFβ signaling pathways.
Across species, BMP signaling has been visualized in embryos using antibodies that specifically recognize the phosphorylated form of the BMP-responsive r-SMAD(s). This approach has been used to observe gradients of BMP signaling activity that correspond with the dorsal/ventral axis in fly, fish, and frog embryos (Dorfman and Shilo, 2001; Plouhinec and De Robertis, 2009; Schohl and Fagotto, 2002; Tucker et al., 2008). In mouse, no graded pSMAD1/5/9 pattern has been reported. Prior to implantation, pSMAD1/5/9 is reportedly detected in all cell types of the embryo at multiple stages (Graham et al., 2014; Reyes de Mochel et al., 2015). After implantation, pSMAD1/5/9 is detected within a subdomain of extraembryonic cells, and not within the embryo itself until it is detected in primordial germ cells and emerging mesoderm during gastrulation (Senft et al., 2019). These observations suggest fundamental differences in the roles of BMP signaling between mammalian and non-mammalian animal embryos, but raise the need for additional, functional lines of evidence.
In mice, individual members of the BMP signaling pathway appear to be dispensable prior to embryonic day 6.5 (E6.5). Knockout of genes encoding the predominant ligand Bmp4 (Lawson et al., 1999; Winnier et al., 1995), the receptors Bmpr2 (Beppu et al., 2000), Bmpr1a (Mishina et al., 1995), Actr1a (Gu et al., 1999), the r-SMADs encoded by Smad1 (Tremblay et al., 2001) and Smad5 (Chang et al., 1999), and the co-Smad Smad4 (Sirard et al., 1998; Yang et al., 1998; Yang et al., 2002) all point to essential roles for BMP signaling in extraembryonic mesoderm, extraembryonic endoderm, and germ cell development. Mechanistically, BMP also interacts with Nodal to pattern the visceral endoderm and to specify distal, and then anterior visceral endoderm, structures required to spatially pattern the embryo and specify the primitive streak (Robertson, 2014; Waldrip et al., 1998; Yamamoto et al., 2009). These events define gastrulation and anterior/posterior axial patterning in mouse, processes which therefore rely on BMP signaling. None of these studies reported that BMP signaling loss-of-function had any effect on development prior to E5.5. However, maternal gene products provided within the oocyte could complicate interpretation of knockout phenotypes resulting from zygotic gene deletion only. Indeed, evidence exists that BMP pathway members are maternally supplied and functional in embryos of other animal species (Das et al., 1998; Faure et al., 2000; Kramer et al., 2002; Miyanaga et al., 2002; Zhang et al., 2020) Finally, mouse embryos are particularly challenging to recover between E4.5 and E6.5 and we lack an in vitro protocol that robustly recapitulates in vivo development during these stages, presenting a barrier to the facile testing of a possible role for BMP signaling during the peri-implantation period.
By contrast, preimplantation embryos are relatively easy to isolate and culture in vitro. Accordingly, several studies have suggested a role for BMP signaling in preimplantation development. Culturing preimplantation embryos in the presence of small-molecule BMP inhibitors led to decreased numbers and cell cycle rate of extraembryonic trophectoderm (TE) and primitive endoderm (PrE) cells, as well as changes in expression of lineage-specific transcription factors, including markers of PrE (SOX17, GATA6), TE (CDX2), and inner cell mass (ICM, OCT4) (Graham et al., 2014; Reyes de Mochel et al., 2015; Stuart et al., 2019). Some of these observations were recapitulated following microinjection of siRNA against Bmp4 or overexpression of dominant-negative forms of Bmpr2 (Graham et al., 2014). Overexpression of dominant-negative Smad4 reportedly phenocopied loss of the upstream signaling components. In principle, these approaches could interfere with the activities of both maternally and zygotically expressed signaling components and thereby achieve more extreme loss of function. However, pSMAD1/5/9 was not examined in these manipulated embryos, so the extent to which these manipulations disrupted BMP signaling is unclear. Moreover, inhibitors are prone to off-target effects, which could further confound interpretation of results (Lowery et al., 2016).
In the present study, we visualize pSMAD1/5/9 in wild-type and embryos in which Bmp4 has been maternally and zygotically deleted, as well as lineage specification and morphogenesis in embryos lacking maternal and zygotic Bmp4 or Smad4 throughout pre-implantation, peri-implantation, and early post-implantation stages. We report that, in contrast to previous studies, BMP signaling is apparently dispensable during mouse preimplantation development. However, we observed that SMAD4-mediated signaling is essential for peri-implantation epiblast morphogenesis shortly after implantation, when it helps attenuate FGF/ERK signaling to enable the timely execution of epiblast morphogenetic events.
Results
Phosphorylated SMAD1/5/9 is first detectable in peri-implantation embryos
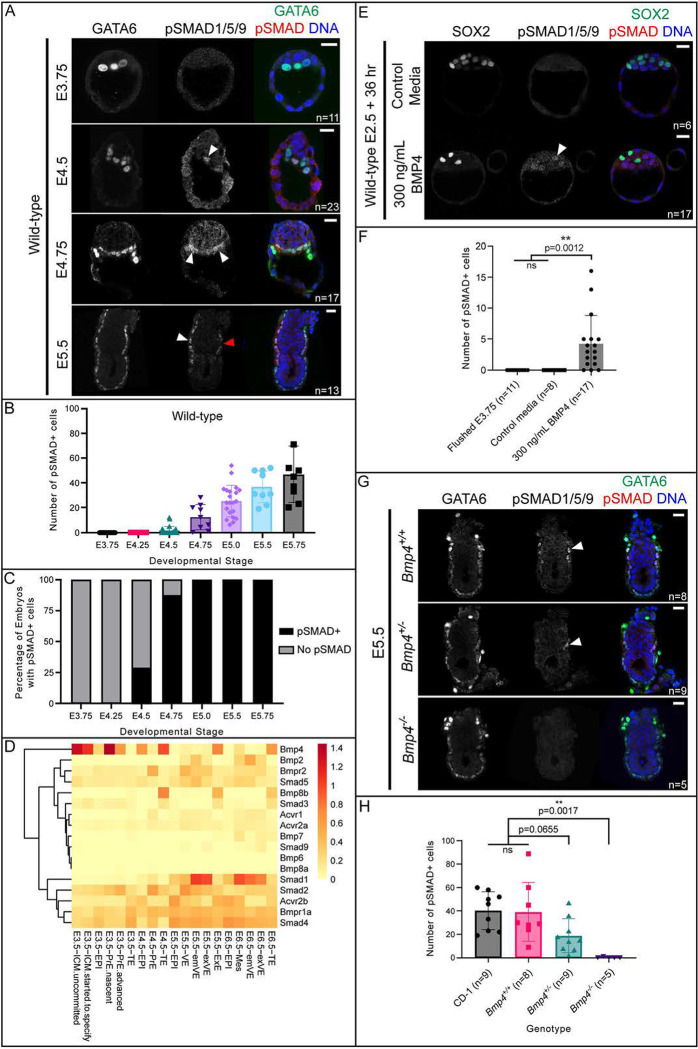
To determine when BMP signaling becomes active in the mouse embryo, we first developed a method to examine the localization of transcription factors SMAD1, 5, and 9, which are phosphorylated in response to ligand/receptor binding (Dijke and Hill, 2004). To achieve this, we used immunofluorescence and an antibody that recognizes phosphorylated SMAD1/5/9 (pSMAD1/5/9, Fig. 1) (Senft et al., 2019; Xu et al., 2019; Yuan et al., 2015). We did not detect pSMAD1/5/9 in preimplantation embryos flushed from uteri between E3.75-E4.25 (Fig 1A, Supp. Fig. 1A). We first observed pSMAD1/5/9 in E4.5 peri-implantation embryos (Fig. 1A), when it was detected in nuclei of a few inner cell mass cells in 29% of embryos examined (Fig. 1B–C). By E4.75, when embryos have undergone implantation, we observed pSMAD1/5/9-positive cells in 87.5% of the embryos evaluated (Fig. 1A–C). Starting at E5.0, we observed pSMAD1/5/9-positive cells within 100% of embryos examined (Fig. 1A–C, Supp. Fig. 1A). The observed pSMAD1/5/9 overlapped with a sub-set of GATA6-expressing primitive endoderm (E4.5 and E4.75) and visceral endoderm (E5.5) cells (Fig. 1A).
Figure 1. BMP signaling becomes active in primitive endoderm at implantation.
A) SMAD1/5/9 phosphorylation (pSMAD1/5/9) in wild-type CD-1 embryos at E3.75, E4.5, E4.75, and E5.5. In all cases, positive pSMAD1/5/9 signal co-localizes with GATA6 as a marker of primitive endoderm and visceral endoderm. B) Quantification of total number of pSMAD1/5/9-positive cells in wild-type embryos in A and Supplemental Figure 1A. C) Quantification of the percentage of embryos from A and Supplemental Figure 1A which display any pSMAD1/5/9-positive cells versus no pSMAD1/5/9-positive cells. D) Heat map of the mean normalized expression of BMP pathway genes from scRNA-seq data from Nowotschin et al., 2019. E) pSMAD1/5/9 in wild-type embryos collected at E2.75 and cultured for 36 hours in media containing 300 ng/mL exogenous BMP4. F) Quantification of the total number of pSMAD1/5/9-positive cells in embryos from E revealed significantly more pSMAD1/5/9-positive cells in BMP4-treated embryos. G) pSMAD1/5/9 staining is absent in Bmp4 z null embryos at E5.5. H) Quantification of total number of pSMAD1/5/9-positive cells in wild-type and Bmp4-null embryos at E5.5 revealed significantly fewer pSMAD1/5/9-positive cells in Bmp4-null embryos. All pairwise comparisons were assessed by analysis of variance (ANOVA) with Tukey’s post-hoc test. White arrowheads indicate positive pSMAD1/5/9 signal. Red arrowhead indicates a GATA6+ cell which does not express pSMAD1/5/9. Scale bars represent 10 μm.
To determine whether the observed pSMAD1/5/9 signal was specific, we treated E5.5 wild-type embryos with LDN-193189 (LDN hereafter) which has been used to disrupt BMP signaling in mouse embryos (Graham et al., 2014; Reyes de Mochel et al., 2015). A concentration of 1 μM LDN was reported as sufficient to inhibit BMP signaling in preimplantation mouse embryos (Reyes de Mochel et al., 2015). However, we found that treatment with 1 μM LDN was highly toxic to embryos (Supp. Fig. 1B). Nevertheless, treatment with 0.25 μM LDN led to complete loss of pSMAD1/5/9 signal in E5.5 embryos (Supp. Fig 1B). Altogether, these observations suggest that BMP signaling becomes active around the time of embryo implantation but is not active during preimplantation stages.
BMP pathway members are present, but largely inactive, prior to implantation
A prior report showed that BMP4 is sufficient to influence gene expression in preimplantation mouse embryos CITE, suggesting that preimplantation embryos can respond to exogenous BMP signals. We therefore examined expression dynamics of genes encoding BMP pathway members during preimplantation stages. We analyzed published single-cell RNA-seq data from mouse embryos at stages E3.5-E6.5 (Nowotschin et al., 2019). At E3.5, many core components of canonical BMP signaling were detectable, including the ligand Bmp4, Type I receptor Bmpr1a, Type II receptors Bmpr2 and Acvr2b, receptor-associated SMAD Smad5, and co-factor SMAD Smad4 (Fig. 1D and Supp. Fig. 2).
Next, we investigated whether pSMAD1/5/9 could be induced in preimplantation embryos treated with exogenous BMP. We cultured compacted 8-cell stage embryos (E2.75) in 300 ng/ml BMP4 for 36 hours to the blastocyst stage (equivalent in cell number to E3.75, as confirmed by cell counts). Although we did not observe pSMAD1/5/9 in any control embryos cultured in unsupplemented medium, we observed low, but detectable levels of pSMAD1/5/9 in 82% (n=14/17) of embryos treated with exogenous BMP4, further supporting it faithful detection of pSMAD1/5/9 by immunofluorescence analysis (Fig. 1E–F). Notably, pSMAD1/5/9 was detected only in the ICM but did not preferentially colocalize with either SOX2-positive epiblast (EPI) or SOX2-negative PrE cells, indicative of normal ICM differentiation. Therefore, we conclude that BMP signaling is not highly active during preimplantation development, but ICM cells are competent to respond to exogenous BMP signals at these stages, consistent with published investigations CITE.
We next evaluated pSMAD1/5/9 in embryos shortly after implantation. Consistent with prior reports (Senft et al., 2019; Yamamoto et al., 2009), we detected pSMAD1/5/9 within a zone of the visceral endoderm (VE) that flanks the extraembryonic ectoderm (EXE) at E5.5 and E5.75 (Fig. 1A, Supp. Fig. 1A). This observation is also consistent with evidence that several key components, including Bmp2, Smad1, Smad5, and Bmpr2, are substantially upregulated around the time of implantation (E4.5-E5.5), particularly in the PrE/VE lineage (Fig. 1D, Supp. Fig 2A–B). Notably, culturing E5.5 embryos in the presence of exogenous BMP4 for 6 hours was sufficient to expand the zone of pSMAD1/5/9 within the VE in a dose-dependent manner (Supp. Fig. 1C). Thus, the availability of ligand could limit the extent of pathway activation, during both pre- and post-implantation stages.
Finally, we evaluated pSMAD1/5/9 in Bmp4-null embryos at E5.5. We were unable to detect pSMAD1/5/9in Bmp4-null embryos, although it was observed at wild-type levels and localization in homozygous wild-type littermate controls (Fig. 1G–H). In Bmp4 heterozygous embryos, we observed an intermediate phenotype where some pSMAD1/5/9 was detectable but trended toward lower numbers of pSMAD1/5/9-positive cells than wild-type (Fig. 1H). This suggests that at E5.5, BMP4 plays a major role in initiating BMP signaling activity in the mouse, and that this function of BMP4 is dose-dependent.
Maternal Bmp4 and Smad4 are not required for development
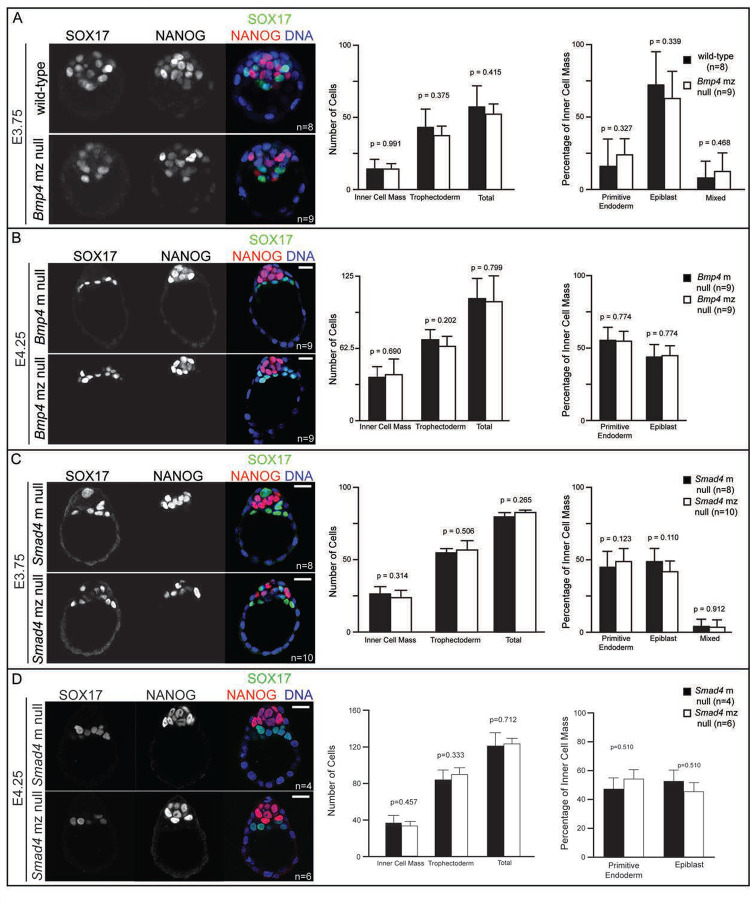
Previous knockout studies of BMP signaling components did not report preimplantation phenotypes (Beppu et al., 2000; Mishina et al., 1995; Sirard et al., 1998; Winnier et al., 1995) However, other groups reported defects in preimplantation lineage specification using pathway inhibitors or microinjection of RNAi or mRNA for dominant-negative overexpression (Graham et al., 2014; Reyes de Mochel et al., 2015; Stuart et al., 2019) One way to reconcile these disparate findings is to invoke a model in which some components of the BMP pathway are maternally imparted to the oocyte and participate in preimplantation development to compensate for previously reported zygotic null mutations. To further investigate this possibility, we examined cell fate specification in embryos lacking both maternal (m) and zygotic (z) Bmp4 or Smad4 using the female germ line-expressed Zp3-Cre (De Vries et al., 2000) in combination with floxed alleles of either Bmp4 or Smad4 (see Supp. Fig. 3A for breeding scheme). RT-qPCR analysis confirmed the absence of detectable Smad4 transcript in Smad4 mz null embryos (Supp. Fig. 3B), as we have observed for many other loci deleted using this approach (Blij et al., 2012; Frum et al., 2013; Frum et al., 2018; Wicklow et al., 2014).
Remarkably, we were able to recover either Bmp4 or Smad4 mz null blastocysts at predicted rates, indicating no requirement for maternal Bmp4 or Smad4 on fertilization or embryo development. Moreover, both Bmp4 and Smad4 mz null embryos exhibited normal morphology, total cell number, and ratio of trophectoderm and ICM cells (Fig 2, Supp. Fig. 3C–E). In addition, the ICM marker Oct4, was detected at normal levels within the ICM, and CDH1 localization strongly suggested that the TE was properly polarized (Supp. Fig. 4). Finally, the expression of EPI and PrE cell fate markers at E3.75, E4.25, and E4.5 (Fig. 2, Supp. Fig. 3C–E) was also unaffected in either Bmp4 or Smad4 mz null embryos at these stages, consistent with normal ICM differentiation. Our observations support the conclusion that canonical BMP signaling does not play a major role in preimplantation development. In a parallel set of experiments, we allowed Smad4 m null embryos to develop to term. Mice lacking m Smad4 were born and developed apparently normally to 4 months old (6/6 mice, one litter). We conclude that maternal Bmp4 and Smad4 are dispensable for development and that neither zygotic gene plays a predominant role prior to implantation.
Figure 2. Maternal and zygotic Smad4 and Bmp4 are dispensable for blastocyst formation and preimplantation cell fate specification.
A) Immunofluorescence for SOX17 and NANOG as respective markers of primitive endoderm (PrE) and epiblast (EPI) in flushed E3.75 wild-type CD-1 embryos and embryos lacking maternal and zygotic Bmp4 (mz null). Quantification did not reveal any significant difference in cell number or cell fate between Bmp4 mz null embryos and controls. “Mixed” indicates co-expression of SOX17 and NANOG. B) Immunofluorescence for SOX17 and NANOG in flushed E4.25 embryos lacking maternal Bmp4 only (m null) and Bmp4 mz null embryos. Quantification did not reveal any significant difference in cell number or cell fate between Bmp4 mz null embryos and controls. “Mixed” indicates co-expression of SOX17 and NANOG. C) Immunofluorescence for SOX17 and NANOG in flushed E3.75 Smad4 m null and Smad4 mz null embryos. Quantification did not reveal any significant difference in cell number or cell fate between Smad4 mz null embryos and controls. D) Immunofluorescence for SOX17 and NANOG in flushed E4.25 Smad4 m null and Smad4 mz null embryos. Quantification did not reveal any significant difference in cell number or cell fate between Smad4 mz null embryos and controls. All pairwise comparisons were assessed by Student’s t-test. Scale bars represent 10 μm.
Smad4 is required for epiblast cavitation at E5.5 in a Bmp4-independent manner
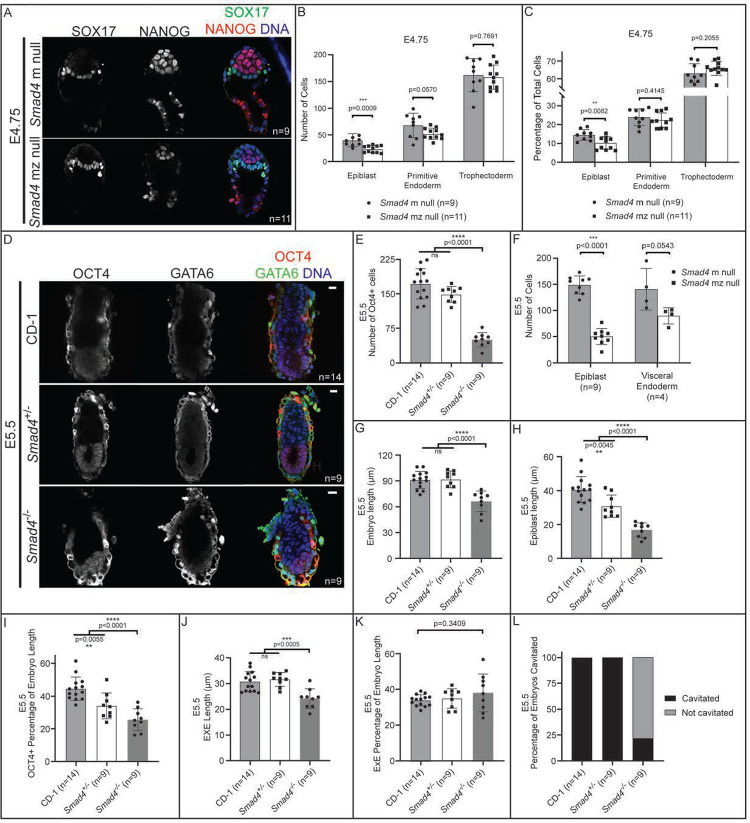
Prior studies mainly focused on characterization of BMP signaling loss of function phenotypes at later stages (>E5.5) (Sirard et al., 1998; Winnier et al., 1995; Yang et al., 1998). However, we first observed BMP signaling activity in most embryos just after implantation at E4.75, prompting us to examine embryos lacking Bmp4 or Smad4 beginning at E4.75. At E4.75, Smad4 null embryos were grossly morphologically normal (Fig. 3A). However, upon close examination, Smad4 null embryos displayed a significant decrease in total cell number (Fig. 3B). We quantified the number of EPI, PrE, and TE cells in these embryos and discovered that the decreased cell number was most pronounced in EPI cells at this stage (Fig. 3B–C).
Figure 3. BMP-independent function of Smad4 is required for post-implantation epiblast organization and maintenance.
A) E4.75 Smad4 mz null embryos stained by immunofluorescence for SOX17 and NANOG. B) Quantification of EPI, PrE, and TE cell numbers from embryos in A revealed a significant decrease in EPI cells in Smad4 mz null embryos when compared to controls. C) Quantification of the EPI, PrE, and TE cells as a percentage of total cell number from embryos in A revealed a significant decrease in EPI percentage in Smad4 mz null embryos. D) E5.5 Smad4−/− embryos stained by immunofluorescence for OCT4 and GATA6 as markers of EPI and VE, respectively. Smad4−/− refers to combined Smad4 z null and Smad4 mz null embryos. E) Quantification of the number of OCT4+ cells in wild-type, Smad4+/−, and Smad4−/− embryos. F) Quantification of EPI and PrE cell numbers from Smad4+/− and Smad4−/− embryos at E5.5 revealed a specific, significant decrease in epiblast cell number in Smad4 mz null embryos when compared to controls (p<0.05 by Student’s t-test). The difference in VE cell numbers was not significant (p>0.05). G) Quantification of the proximal-distal length wild-type, Smad4+/−, and Smad4−/− embryos at E5.5. H) Quantification of the proximal-distal length of the EPI of wild-type, Smad4+/−, and Smad4−/− embryos at E5.5. I) Quantification of the proximal-distal length of the EPI as a percentage of total length of wild-type, Smad4+/−, and Smad4−/− embryos at E5.5. J) Quantification of the proximal-distal length of the EXE of wild-type, Smad4+/−, and Smad4−/− embryos at E5.5. K) Quantification of the proximal-distal length of the EXE as a percentage of total length of wild-type, Smad4+/−, and Smad4−/− embryos at E5.5. L) Quantification of the proportion of Smad4+/− and Smad4−/− embryos with a proamniotic cavity at E5.5. Comparisons in B, C, F were assessed by Student’s t-test. Comparisons in E, G-K were assessed by analysis of variance (ANOVA) with Tukey’s post-hoc test.
By E5.5, Smad4 null embryos were visibly reduced in size and all displayed disorganization in EPI, VE, and EXE compartments, as expected from studies performed at E6.5 (Sirard et al., 1998; Yang et al., 1998) (Fig. 3D). Strikingly, the epiblast was greatly reduced in cell number (Fig. 3D–F) relative to controls, and had not yet cavitated, as the proamniotic cavity was not present among most (>80%, n=7/9) Smad4 null embryos examined (Fig. 3D, 3L). Notably, relative to extraembryonic lineages, the number of EPI cells was disproportionately decreased in Smad4 null embryos since the epiblast length was decreased even when normalized to proximal-distal embryo length (Fig. 3G–I, Supp. Fig. 5A). By contrast, the size of the EXE was appropriately scaled to the reduced size of Smad4 null embryos at E5.5 (Fig. 3J–K). This suggests that Smad4 is not only required for general embryonic growth, but also specifically required for epiblast growth relative to total embryo size. Notably, these phenotypes were not observed in E5.5 Bmp4-null embryos, which did not differ from wild-type in morphology, embryo size, or EPI or PrE cell number (Supp. Fig. 5B–E), suggesting that alternative signaling pathways feed into SMAD4-regulation of epiblast growth and morphogenesis at this stage.
Epiblast cavitation requires SMAD4-dependent inhibition of FGF/ERK signaling
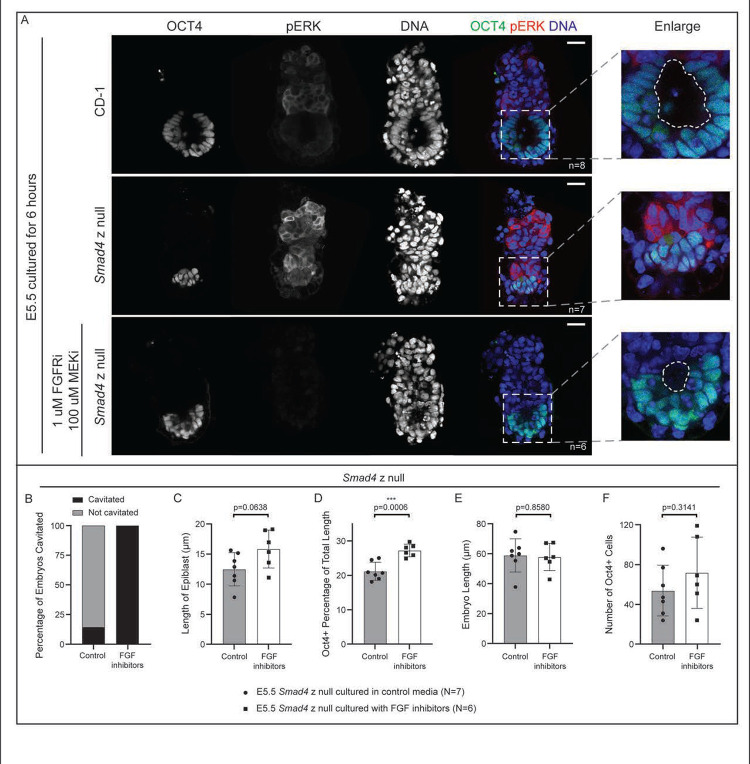
Having discovered that Smad4 is required for epiblast cavitation and growth at E5.5, we began to investigate the mechanism underlying this role. We were struck by the observation that the EXE appeared disproportionately large, relative to the size of the EPI in E5.5 Smad4 null embryos. To confirm the identity of the EXE cells, we examined markers of EXE, including phosphorylated ERK (pERK), which is elevated within the EXE (Corson et al., 2003). In Smad4 null embryos, we observed pERK throughout putative EXE, consistent with their identity as EXE cells. However, we observed dramatically elevated levels of pERK within this region (Fig. 4A). Since pERK in the EXE is known to be dependent on signaling by the Fibroblast Growth Factor (FGF) pathway (Corson et al., 2003), we therefore hypothesized that increased pERK could be due to elevated FGF signaling in Smad4 null embryos.
Figure 4. Inhibition of FGF signaling partially rescues epiblast cavitation in E5.5 Smad4 null embryos.
A) Wild-type and Smad4 z null embryos collected at E5.5 and cultured for 6 hours after dissection in control media or media containing FGFR/MEK inhibitors (see Methods), then stained by immunofluorescence for OCT4 and phosphorylated ERK (pERK). Dashed line in enlargement denotes the proamniotic cavity. B) Quantification of the proportion of treated and untreated Smad4−/− embryos with a proamniotic cavity at E5.5. C) Quantification of proximal-distal length of the EPI in treated and untreated E5.5 Smad4−/− embryos. D) Quantification of proximal-distal length of the EPI as a proportion of total length in treated and untreated E5.5 Smad4−/− embryos. E) Quantification of proximal-distal length in treated and untreated E5.5 Smad4−/− embryos. F) Quantification of OCT4-positive cell number in treated and untreated E5.5 Smad4−/− embryos.
To test this hypothesis, we used a previously-published protocol to inhibit FGF signaling in embryos (Yamanaka et al., 2010), which effectively eliminated pERK in control embryos (Supp. 6A). Notably, inhibition of FGF signaling partially rescued epiblast defects in E5.5 Smad4 null embryos (Fig. 4A). We observed a significant increase in epiblast scaling, as well as increased rates of cavitation (Fig. 4B–D). However, FGF inhibitor treatment did not rescue the overall growth restriction of Smad4-null embryos (Fig. 4E), nor was the number of OCT4-positive cells restored (Fig. 4F). These observations are consistent with SMAD4 repressing FGF signaling in the EXE during early post-implantation stages as a critical regulator of epiblast morphogenesis. Our observations also suggest that additional pathways regulate embryo growth downstream of SMAD4.
To determine whether elevated FGF signaling is sufficient to antagonize cavitation, we treated embryos with exogenous FGF4. In wild-type E5.5 embryos treated with exogenous 1 μg/mL FGF4, we observed elevated levels of pERK within the EXE and ectoplacental cone (Supp. Fig. 6A). However, we observed no impact on epiblast cavitation or epiblast size following this treatment (Supp. Fig. 6A–D). Altogether, these data suggest that pERK/FGF signaling antagonizes EPI cavitation, but upregulation of pERK alone is insufficient to induce cavitation defects in wild-type embryos, at least under the conditions tested here.
Discussion
Despite being a highly conserved developmental signaling pathway, the role of BMP signaling in regulating peri-implantation mammalian development has not been fully elucidated. In part, this may be due to the technical challenges surrounding studying mammalian embryos at pre- and peri-implantation stages of development. Our study undertook a detailed examination of molecular markers of BMP signaling activity and the phenotypes of maternal-zygotic genetic knockout models of BMP signaling to identify the earliest role of BMP signaling in development. Although we failed to detect a role for BMP signaling in preimplantation embryos, we identified a novel role for Smad4 in epiblast organization and cavitation at early post-implantation stages.
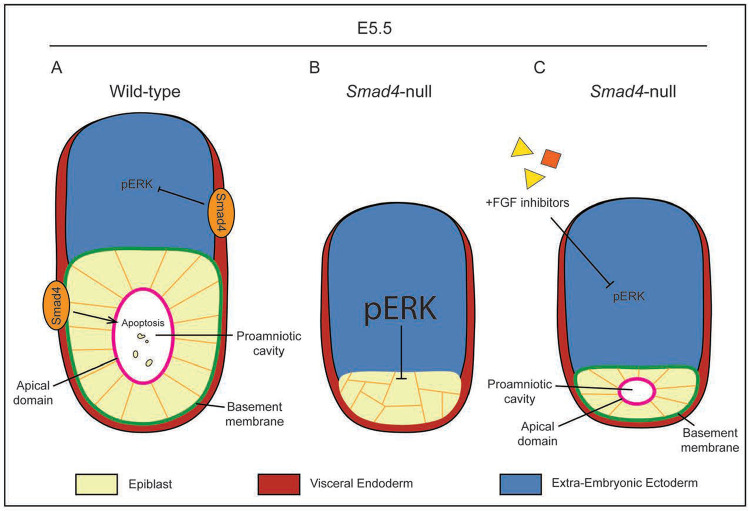
We report a novel requirement for Smad4 in post-implantation development. Previous descriptions of Smad4-null embryos report growth restriction and disorganized VE beginning at E5.5, with embryonic lethality by E8.5 (Chu et al., 2004; Sirard et al., 1998; Yang et al., 1998). Our results are consistent with these findings, but also uncover a previously unappreciated defect specific to the epiblast. The proportion of EPI cells are decreased starting at E4.75, suggesting that in the absence of Smad4 the number of EPI cells does not scale with embryo growth. The structure of the epiblast is also disrupted in these embryos, as Smad4-null embryos had largely disorganized epiblasts which failed to form a proamniotic cavity. It is unlikely that this defect is cell-autonomous, as embryos with an epiblast-specific knockout of Smad4 do not display an observable defect until after gastrulation (Chu et al., 2004). Rather, it is likely that Smad4 is required in the visceral endoderm to produce a signal that promotes EPI development non-cell-autonomously (Fig. 5A). This model is supported by the finding that VE-specific Smad4-knockout embryos fail to gastrulate properly and more closely resemble whole-body Smad4-null embryos (Li et al., 2010).
Figure 5. FGF inhibition rescues rosette formation but not embryo growth in Smad4-null embryos.
A) In wild-type embryos, SMAD4 in the visceral endoderm promotes lumen formation in the epiblast by regulating a pro-apoptotic signal. Separately, SMAD4 also inhibits ERK phosphorylation in the EXE, which allows for polarization and rosette formation in the epiblast. B) In Smad4-null embryos, pERK is upregulated, causing an ectopic increase in pERK and preventing epiblast polarization. The pro-apoptotic signal is also lost. The combination of these two factors lead to epiblast disorganization and a failure to cavitate. C) Treatment with FGF inhibitors prevents upregulation of pERK in Smad4-null embryos. Repressed pERK levels allow epiblast polarization to proceed, resulting in a small proamniotic cavity.
Our observations are consistent with prior observation that BMP signaling is necessary and sufficient for cavitation of embryoid bodies (Coucouvanis and Martin, 1999). In this context, BMP2 and BMP4 may be functionally redundant, and could explain why we observed normal cavitation in E5.5 Bmp4-null embryos. Alternatively, SMAD4 could regulate epiblast morphogenesis through another TGF-β pathway. Knockout of Nodal has been shown to decrease embryo size and expression of Oct4 mRNA at early post-implantation stages (Brennan J. et al., 2001; Mesnard et al., 2006), but Nodal-null embryos cavitate normally and expression of OCT4 protein is apparently unaffected at E5.5 (Senft et al., 2019). As the cavitation defect and loss of OCT4-positive cells are more severe in Smad4-null embryos than either Bmp4- or Nodal-null models alone, it suggests that Smad4 may regulate epiblast morphogenesis through a combination of TGFβ pathways.
Our data also suggests that SMAD promotes cavitation by attenuating pERK levels. Inhibition of FGF/MAPK signaling rescued cavitation in Smad4-null embryos, even though the epiblast remained disproportionately small. This observation allows us to propose that cavitation is not dependent on epiblast size, but rather the embryo signaling environment. Our data suggest that ectopic upregulation of pERK resulting from loss of Smad4 is detrimental to epiblast cavitation; however, the mechanisms of how these two pathways interact with one another and regulate cavitation remain unclear. SMAD4 could regulate several processes associated with epiblast cavitation, including epiblast cell apoptosis or epiblast cell polarization and lumenogenesis (Bedzhov and Zernicka-Goetz, 2014; Coucouvanis and Martin, 1995; Coucouvanis and Martin, 1999; Halimi et al., 2022) (Fig 5B–C). Alternatively, SMAD4 could promote epiblast maturation, defined here as the transition from a naïve to primed state which normally occurs in epiblast cells between preimplantation and post-implantation stages (Boroviak et al., 2015; Nichols and Smith, 2009). Epiblast maturation has been shown to be critical in formation of proamniotic cavity (Carbognin et al., 2023; Shahbazi et al., 2017). Other mechanisms are also proposed to play a role in embryonic cavitation such as apical domain maintenance (Meng et al., 2017), tight junction formation (Chan et al., 2019), cellular adhesion through ECM interactions (Liang et al., 2005; Sakai et al., 2003), and establishment of an osmotic gradient (Dumortier et al., 2019). SMAD4 and pERK may regulate any or a combination of these factors. We note that experimental elevation of pERK was not sufficient to inhibit cavitation in wild-type embryos, at least under the conditions tested here. This observation could suggest that pERK regulates expression of components of the cavitation process that are not themselves rate-limiting. Further studies will be needed to interrogate the interaction of these pathways further, to identify the precise regulatory mechanisms governing post-implantation epiblast morphogenesis downstream of SMAD4 and pERK.
Materials and Methods
scRNA-seq Analysis
Single-cell RNA-seq data generated by Nowotschin et. al. was used to analyze the expression of TGFβ genes in mouse E3.5, E4.5, E5.5, and E6.5 blastocysts (Nowotschin et al., 2019) (Nowotschin et al., 2019). The analysis was completed using R v4.1.0 with tools from Seurat v4.3.0 (R-core Team 2021) (Hao et al., 2021). We normalized the UMI counts using SCTransform and cells were visualized in 2D space using UMAP performed on the first 30 principal components (Choudhary and Satija, 2022; Hafemeister and Satija, 2019). After excluding TGFβ genes expressed in <10 cells, we used Seurat’s FindAllMarkers function with the Wilcoxon rank-sum test to identify TGFβ genes enriched in each cell type versus all other cells. The p-values were corrected for multiple comparisons using the Bonferroni method. Genes with p-adj<0.01 and average log2 fold change<0.25 were considered cluster enriched. Heatmaps were generated using the pheatmap (v 1.0.12) after averaging the normalized expression for each gene in each cell type.
Mouse Strains and Genotyping
All animal research was conducted in accordance with the guidelines of the Michigan State University Institutional Animal Care and Use Committee. Wild type embryos were derived from CD-1 mice (Charles River). The following alleles were used in this study and maintained in a CD-1 background: Bmp4tm1Jfm/J (Liu et al., 2004); Smad4tm2.1Cxd/J (Yang et l., 2002); Tg(Zp3-cre)93Knw (de Vries et al., 2000). Null alleles were generated by breeding dams carrying homozygous floxed alleles and the Zp3Cre allele to CD-1 males. Mouse genotypes were determined by PCR using genomic DNA extracted using the REDExtract-N-Amp kit (Sigma XNAT) according to the manufacturer’s protocol. Embryo genomic DNA was extracted using the same kit scaled to 10 μL total volume. Genomic extracts (1–2 μL) were then subjected to PCR using allele-specific primers (see Table 3).
Table 3.
Allele-specific primers for PCR genotyping
| Allele | Forward Primer | Reverse Primer |
|---|---|---|
| Bmp4WT, Bmp4fl | GAGCTAAGTTTTGCTGGTTTGC | GCCCATGAGCTTTTCTGAGA |
| Bmp4del | GCTAAGTTTTGCTGGTTTGC | TGACTAGGGGAGGAGTAGAAGGTG |
| Smad4WT, Smad4fl | TAAGAGCCACAGGGTCAAGC | TTCCAGGAAAAACAGGGCTA |
| Smad4del | TAAGAGCCACAGGGTCAAGC | GACCCAAACGTCACCTTCAG |
| Cre | CTAGGCCACAGAATTGAAAGATCT | GTAGGTGGAAATTCTAGCATCATCC |
| Zp3Cre | CGAGATTGAGGGAAGCAGAG | CAGGTTCTTGCGAACCTCAT |
Embryo Collection and Culture
Mice were maintained on a 12 hr light/dark cycle. Preimplantation (E2.5-E4.5) embryos were collected by flushing the oviduct or uterus with M2 medium (Sigma M7167). Post-implantation (E4.75-E6.5) embryos were collected by dissecting the embryos from the decidua in ice-cold PBS containing 1% FBS (HyClone SH30396.02) or Bovine Serum Albumin (BSA, Sigma A7888). During embryo collection, dissected embryos were held in warm M2 media. For embryo culture, KSOM medium (Millipore MR-121-D) was equilibrated overnight prior to embryo collection. Where indicated, the following were included in the culture medium: 1 μM or 0.25 μM LDN-193189 in DMSO (Stemgent 04-0074-02); 1 μg/mL recombinant FGF4 in PBS with 0.1% BSA (R&D 235-F4); 1 μg/mL heparin (Sigma H3149); 100 ng/mL recombinant BMP4 in 4 mM HCl (R&D 314-BP); 1 μM PD173074 in DMSO (Selleckchem S1264); 5 μM PD0325901 in DMSO (Stemgent 04–0006); or 0.2% DMSO (New England BioLabs B0515A) as control. Embryos were cultured at 37°C in a 5% CO2 incubator under light mineral oil (Millipore ES-005-C).
Real-time PCR of Oocytes
Smad4 expression levels in oocytes were assessed by real-time PCR as previously described (Blij et al, 2012). Smad4 levels were assessed in oocytes from three wild-type and three Smad4 maternal null females. Oocytes collected from each female were pooled for mRNA extraction and cDNA synthesis. RT-PCR was performed in quadruplicate technical replicates for each cDNA sample. Primers were (5′−3′): Actb, CTGAACCCTAAGGCCAACC and CCAGAGGCATACAGGGACAG; Smad4 (wild-type allele) CGCGGTCTTTGTACAGAGTTA and ACACTGCCGCAGATCAAAG; Smad4 (deleted allele), CACAGGACAGAAGCGATTGA and CCAAACGTCACCTTCACCTT.
Immunofluorescence and Confocal Microscopy
Preimplantation embryos (E2.5-E4.75) were fixed with 4% formaldehyde (Polysciences 04018) for 10 min, permeabilized with 0.5% Triton X-100 (Sigma Aldrich X100) for 30 min, and then blocked with blocking solution (10% Fetal Bovine Serum (HyClone SH30396.02), 0.1% Triton X-100) overnight at 4°C. Embryos were incubated with primary antibody overnight at 4°C. The next day, embryos were washed in blocking solution for 30 min, incubated in secondary antibody diluted in blocking solution for 1 hr, washed in blocking solution for 30 min, then stained with nuclear stain diluted in block for 10 min or overnight.
Post-implantation embryos (E5.0-E5.75) were fixed with 4% formaldehyde for 1 hr, washed 3 times in 0.1% Tween-20 (Sigma Aldrich P9416), permeabilized for 4 hrs in 0.5% Triton X-100, and then blocked with blocking solution (3% BSA (Sigma Aldrich A7888); 0.3% Triton X-100 in PBS) overnight at 4°C. Embryos were incubated with primary antibody overnight at 4°C. The next day, embryos were washed three times in 0.1% Tween-20 for 5 min, then incubated in secondary antibody diluted in blocking solution overnight. The following day embryos were washed three times in 0.1% Tween-20 for 5 min, then stained with nuclear stain diluted in block for 10 min or overnight.
All embryos (preimplantation or post-implantation) which used antibodies against pSMAD1/5/9 were fixed with 4% formaldehyde for 1 hr, methanol dehydration-rehydration series (25%, 50%, 75%, 100%) for 5 min each, washed three times in freshly-made 1% Triton X-100 for 10 min, washed 20 min in ice-cold acetone at −20°C, washed three times in freshly-made 1% Triton X-100 for 10 min, then then blocked with blocking solution (10% Fetal Bovine Serum, 0.1% Triton X-100 in PBS) overnight at 4°C. Embryos were incubated with primary antibody overnight at 4°C. The next day, embryos were washed three times in freshly-made 0.1% Triton X-100 for 10 min, incubated in secondary antibody diluted in blocking solution for 2 hrs, washed three times in freshly-made 0.1% Triton X-100 for 10 min, then stained with nuclear stain diluted in blocking solution for 10 min or overnight.
All embryos (preimplantation or post-implantation) which used antibodies against pERK were fixed with 4% formaldehyde for 1 hr, washed three times for 5 min in PBS, washed 20 min in ice-cold methanol at −20°C, permeabilized 30 min in 0.1% Tween-20, then blocked with blocking solution (3% Bovine Serum Albumin; 0.3% Triton X-100 in PBS) overnight at 4°C. Embryos were incubated with primary antibody overnight at 4°C. The next day, embryos were washed three times in PBS for 5 min, incubated in secondary antibody diluted in blocking solution for 2 hrs, washed three times in PBS for 5 min, then stained with nuclear stain diluted in block for 10 min or overnight. All solutions contained HALT protease inhibitor (Thermo Scientific 78430) and PhosSTOP phosphatase inhibitor (Roche 04906837001) diluted 1:500.
Antibodies used are listed in Table 2. Embryos were imaged using an Olympus FluoView FV1000 Confocal Laser Scanning Microscope system with 60X PlanApoN oil (NA 1.42) objective. For each embryo, z-stacks were collected, with 5 μm intervals between optical sections. All embryos were imaged prior to knowledge of their genotypes.
Table 2.
Antibody Table
| Antibody | Source or reference(s) | Identifiers | Dilution | Additional Information |
|---|---|---|---|---|
| Goat-anti-hGATA6 | R&D Systems | AF1700 | 1.100 | |
| Goat-anti-GATA4 | Santa Cruz Biotechnology | sc1237 | 1.2000 | |
| Goat-anti-SOX17 | R&D Systems | AF1924 | 1.2000 | |
| Mouse-anti-CDX2 | Abcam | CDX-88 | 1.500 | |
| Rabbit-anti-CDX2 | Abcam | ab76541 | 1.200 | |
| Mouse-anti-OCT4 | Santa Cruz Biotechnology | 5–5279 | 1.100 | |
| Rabbit-anti-NANOG | Reprocell | RCAB002P-F | 1:400 | |
| Goat-anti-SOX2 | Neuromics | GT15098 | 1.2000 | |
| Rat-anti-CDH1 | Sigma | U3254 | 1.500 | |
| Rabbit-anti-Phospho-Smad1 (Ser463/465)/Smad5 (Ser463/465)/Smad9 (Ser465/467) | Cell Signaling | 13820 | 1.50 | |
| Rabbit-anti-Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) | Cell Signaling | 9101 | 1.100 | |
| Donkey-anti-goat IgG | Invitrogen | A-11055 | 1:400 | Alexa488 |
| Donkey-anti-mouse IgG | Jackson Laboratories | 715–165-150 | 1:400 | Cy3 |
| Donkey-anti-rabbit IgG | Jackson Laboratories | 711–165-152 | 1:400 | Cy3 |
| Goat-anti-rabbit IgG | Invitrogen | A-11034 | 1:400 | Alexa488 |
| Goat-anti-mouse IgG | Invitrogen | A-11029 | 1:400 | Alexa488 |
| DRAQ5 | Cell Signaling | 4084 | 1:400 |
Embryo Analysis
For each embryo, z-stacks were analyzed using Fiji (ImageJ), which enabled the labeling, based on DNA stain, of all individual cell nuclei. Using this label to identify individual cells, each cell in each embryo was then assigned to relevant phenotypic categories, without knowledge of embryo genotype. Phenotypic categories included marker expression (e.g., OCT4 positive or negative) and marker localization (e.g., pSMAD1/5/9 nuclear, absent, or unlocalized). Statistical analysis was performed using GraphPad Prism (v. 9.5.1). Figure images were assembled using Adobe Illustrator.
Supplementary Material
Table 1.
Animal Resources Table
| Allele | Common Allele Name | Strain, strain background (species) | Source or Reference | Identifier(s) | Additional Information |
|---|---|---|---|---|---|
| CD-1 | CD-1 | CD-1 (Mus musculus) | Charles River Laboratories | Crl:CD1(ICR), Strain code: 022 | Wild-type |
| Tg(Zp3-cre)93Knw/J | Zp3Cre | CD-1 (M. musculus) | De Vries et al, 2000 (PMID: 10686600) | RRID:IMSR_JAX:003651 MGI:J:67903 | Presence of this allele assessed by genotyping for Cre and Zp3Cre |
| Bmp4tm1Jfm/J | Bmp4fl | CD-1 (M. musculus) | Liu et al, 2004 (PMID: 15070745; PMCID: PMC384774) | RRID:IMSR_JAX:016878 MGI:J:89237 | Bmp4del or Bmp4 null refers to recombined allele |
| Smad4tm2.1Cxd/J | Smad4fl | CD-1 (M. musculus) | Yang et al, 2002 (PMID: 11857783) | RRID:IMSR_JAX:017462 MGI:J:75140 | Smad4del or Smad4 null refers to recombined allele |
Acknowledgements
We thank the lab of Dr. David Arnosti for their generous loan of laboratory equipment. We also thank Barbara Makela and Ella Markley for technical support.
Funding
REK and MAS were supported by National Institutes of Health (NIH) Award T32 HD087166. This study was supported by NIH R35 GM131759 to AR. Work in the laboratory of KKN was supported by the Wellcome (221856/Z/20/Z) and by the Francis Crick Institute (FC001120).
Diversity and Inclusion Statement
The authors wholeheartedly support all efforts to increase the inclusion of scientists from underrepresented backgrounds (including, but not limited to, women, LGBTQIA+, people of color, Indigenous people, neurodivergent people, people with disabilities, and people from disadvantaged backgrounds) in developmental biology and related careers. This manuscript reflects the efforts of authors who identify as members of several of these groups. The authors believe that diverse perspectives are essential for scientific excellence and innovation, yet acknowledge the continued existence of systemic barriers to success for scientists of underrepresented and marginalized communities. We support the development of initiatives to address disparities and biases in scientific publishing and encourage further efforts to implement inclusive practices.
Footnotes
Competing interests
The authors declare no competing interests.
References
- Bedzhov I. and Zernicka-Goetz M. (2014). Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 156, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beppu H., Kawabata M., Hamamoto T., Chytil A., Minowa O., Noda T. and Miyazono K. (2000). BMP type II receptor is required for gastrulation and early development of mouse embryos. Developmental Biology 221, 249–258. [DOI] [PubMed] [Google Scholar]
- Blij S., Frum T., Akyol A., Fearon E. and Ralston A. (2012). Maternal Cdx2 is dispensable for mouse development. Development (Cambridge) 139, 3969–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Lombard P., Okahara J., Behr R., Sasaki E., Nichols J., Smith A. and Bertone P. (2015). Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Developmental Cell 35, 366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J., Lu C.C., Norris D. P., Rodriguez T.A., Beddington R.S.P. and Robertson E.J. (2001). Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411, 965–969. [DOI] [PubMed] [Google Scholar]
- Carbognin E., Carlini V., Panariello F., Chieregato M., Guerzoni E., Benvegnù D., Perrera V., Malucelli C., Cesana M., Grimaldi A., et al. (2023). Esrrb guides naive pluripotent cells through the formative transcriptional programme. Nature Cell Biology 25, 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. J., Costanzo M., Ruiz-Herrero T., Mönke G., Petrie R. J., Bergert M., Diz-Muñoz A., Mahadevan L. and Hiiragi T. (2019). Hydraulic control of mammalian embryo size and cell fate. Nature 571, 112–116. [DOI] [PubMed] [Google Scholar]
- Chang C. (2016). Agonists and Antagonists of TGF-β Family Ligands. Cold Spring Harb Perspect Biol 8, a021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Huylebroeck D., Verschueren K., Guo Q., Matzuk M. M. and Zwijsen A. (1999). Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development 126, 1631–1642. [DOI] [PubMed] [Google Scholar]
- Choudhary S. and Satija R. (2022). Comparison and evaluation of statistical error models for scRNA-seq. Genome Biology 23, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G. C., Dunn N. R., Anderson D. C., Oxburgh L. and Robertson E. J. (2004). Differential requirements for Smad4 in TGF -dependent patterning of the early mouse embryo. Development 131, 3501–3512. [DOI] [PubMed] [Google Scholar]
- Corson L. B., Yamanaka Y., Venus Lai K. M. and Rossant J. (2003). Spatial and temporal patterns of ERK signalling during mouse embryogenesis. Development 130, 4527–4537. [DOI] [PubMed] [Google Scholar]
- Coucouvanis E. and Martin G. R. (1995). Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83, 279–287. [DOI] [PubMed] [Google Scholar]
- Coucouvanis E. and Martin G. R. (1999). BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development 126, 535–546. [DOI] [PubMed] [Google Scholar]
- Das P., Maduzia L., L., Wang H., L. Finelli A., Cho S.-H., Smith M. M. and Padgett R. W. (1998). The Drosophila gene Medea demonstrates the requirement for different classes of Smads in dpp signaling. Development 125, 1519–1528. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M. and Sasai Y. (1996). A common plan for dorsoventral patterning in Bilateria. Nature 380, 37–40. [DOI] [PubMed] [Google Scholar]
- De Vries W. N., Binns L. T., Fancher K. S., Dean J., Moore R., Kemler R. and Knowles B. B. (2000). Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis 26, 110–112. [PubMed] [Google Scholar]
- Dijke P. ten and Hill C. S. (2004). New insights into TGF-β–Smad signalling. Trends in Biochemical Sciences 29, 265–273. [DOI] [PubMed] [Google Scholar]
- Dorfman R. and Shilo B. Z. (2001). Biphasic activation of the BMP pathway patterns the Drosophila embryonic dorsal region. Development 128, 965–972. [DOI] [PubMed] [Google Scholar]
- Dumortier J. G., Le Verge-Serandour M., Tortorelli A. F., Mielke A., De Plater L., Turlier H. and Maître J. L. (2019). Hydraulic fracturing and active coarsening position the lumen of the mouse blastocyst. Science 365, 465–468. [DOI] [PubMed] [Google Scholar]
- Faure S., Lee M. A., Keller T., Dijke P. ten and Whitman M. (2000). Endogenous patterns of TGFβ superfamily signaling during early Xenopus development. Development 127, 2917–2931. [DOI] [PubMed] [Google Scholar]
- Frum T., Halbisen M. A., Wang C., Amiri H., Robson P. and Ralston A. (2013). Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev Cell 25, 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum T., Murphy T. M. and Ralston A. (2018). HIPPO signaling resolves embryonic cell fate conflicts during establishment of pluripotency in vivo. eLife 7, e42298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham S. J., Wicher K. B., Jedrusik A., Guo G., Herath W., Robson P. and Zernicka-Goetz M. (2014). BMP signalling regulates the pre-implantation development of extraembryonic cell lineages in the mouse embryo. Nat Commun 5, 5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Reynolds E. M., Song J., Lei H., Feijen A., Yu L., He W., MacLaughlin D. T., Van Den Eijnden-Van Raaij J., Donahoe P. K., et al. (1999). The type I serine/threonine kinase receptor ActRIA (ALK2) is required for gastrulation of the mouse embryo. Development 126, 2551–2561. [DOI] [PubMed] [Google Scholar]
- Hafemeister C. and Satija R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology 20, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halimi R., Levin-Zaidman S., Levin-Salomon V., Bialik S. and Kimchi A. (2022). Epiblast fragmentation by shedding—a novel mechanism to eliminate cells in post-implantation mouse embryos. Cell Death and Differentiation 29, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Hao S., Andersen-Nissen E., Mauck W. M., Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S. (2016). Transcriptional Control by the SMADs. Cold Spring Harb Perspect Biol 8, a022079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer C., Mayr T., Nowak M., Schumacher J., Runke G., Bauer H., Wagner D. S., Schmid B., Imai Y., Talbot W. S., et al. (2002). Maternally Supplied Smad5 Is Required for Ventral Specification in Zebrafish Embryos Prior to Zygotic Bmp Signaling. Developmental Biology 250, 263–279. [PubMed] [Google Scholar]
- Lawson K. A., Dunn N. R., Roelen B. A. J., Zeinstra L. M., Davis A. M., Wright C. V. E., Korving J. P. W. F. M. and Hogan B. L. M. (1999). Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13, 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li Y.-P., Fu X.-Y. and Deng C. X. (2010). Anterior Visceral Endoderm SMAD4 Signaling Specifies Anterior Embryonic Patterning and Head Induction in Mice. International Journal of Biological Sciences 6, 569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zhou Q., Li X., Sun Y., Lu M., Dalton N., Ross J. and Chen J. (2005). PINCH1 Plays an Essential Role in Early Murine Embryonic Development but Is Dispensable in Ventricular Cardiomyocytes. Molecular and Cellular Biology 25, 3056–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery J. W., Brookshire B. and Rosen V. (2016). A survey of strategies to modulate the bone morphogenetic protein signaling pathway: Current and future perspectives. Stem Cells International 2016,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. and Sheppard D. (2023). TGF-β signaling in health and disease. Cell 186, 4007–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Cai K. Q., Moore R., Tao W., Tse J. D., Smith E. R. and Xu X. X. (2017). Pten facilitates epiblast epithelial polarization and proamniotic lumen formation in early mouse embryos. Developmental Dynamics 246, 517–530. [DOI] [PubMed] [Google Scholar]
- Mesnard D., Guzman-Ayala M. and Constam D. B. (2006). Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development 133, 2497–2505. [DOI] [PubMed] [Google Scholar]
- Mishina Y., Suzuki A., Ueno N. and Behringer R. R. (1995). Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes and Development 9, 3027–3037. [DOI] [PubMed] [Google Scholar]
- Miyanaga Y., Torregroza I. and Evans T. (2002). A Maternal Smad Protein Regulates Early Embryonic Apoptosis in Xenopus laevis. Mol Cell Biol 22, 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J. and Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487–492. [DOI] [PubMed] [Google Scholar]
- Nowotschin S., Setty M., Kuo Y.-Y., Liu V., Garg V., Sharma R., Simon C. S., Saiz N., Gardner R., Boutet S. C., et al. (2019). The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor M. B., Umulis D., Othmer H. G. and Blair S. S. (2006). Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development 133, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouhinec J.-L. and De Robertis E. M. (2009). Systems Biology of the Self-regulating Morphogenetic Gradient of the Xenopus Gastrula. Cold Spring Harb Perspect Biol 1, a001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes de Mochel N. S., Luong M., Chiang M., Javier A. L., Luu E., Toshihiko F., MacGregor G. R., Cinquin O. and Cho K. W. (2015). BMP signaling is required for cell cleavage in preimplantation-mouse embryos. Dev Biol 397, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson E. J. (2014). Dose-dependent nodal/smad signals pattern the early mouse embryo. Seminars in Cell and Developmental Biology 32, 73–79. [DOI] [PubMed] [Google Scholar]
- Sakai T., Li S., Docheva D., Grashoff C., Sakai K., Kostka G., Braun A., Pfeifer A., Yurchenco P. D. and Fässler R. (2003). Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes and Development 17, 926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schohl A. and Fagotto F. (2002). β-catenin, MAPK and Smad signaling during early Xenopus development. Development 129, 37–52. [DOI] [PubMed] [Google Scholar]
- Senft A. D., Bikoff E. K., Robertson E. J. and Costello I. (2019). Genetic dissection of Nodal and Bmp signalling requirements during primordial germ cell development in mouse. Nature Communications 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M. N., Scialdone A., Skorupska N., Weberling A., Recher G., Zhu M., Jedrusik A., Devito L. G., Noli L., MacAulay I. C., et al. (2017). Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature 552, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard C., de la Pompa J. L., Elia A., Itie A., Mirtsos C., Cheung A., Hahn S., Wakeham A., Schwartz L., Kern S. E., et al. (1998). The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes & development 12, 107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart H. T., Stirparo G. G., Lohoff T., Bates L. E., Kinoshita M., Lim C. Y., Sousa E. J., Maskalenka K., Radzisheuskaya A., Malcolm A. A., et al. (2019). Distinct Molecular Trajectories Converge to Induce Naive Pluripotency. Cell Stem Cell 25, 388–406.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay K. D., Dunn N. R. and Robertson E. J. (2001). Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 128, 3609–3621. [DOI] [PubMed] [Google Scholar]
- Tucker J. A., Mintzer K. A. and Mullins M. C. (2008). The BMP Signaling Gradient Patterns Dorsoventral Tissues in a Temporally Progressive Manner along the Anteroposterior Axis. Developmental Cell 14, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip W. R., Bikoff E. K., Hoodless P. A., Wrana J. L. and Robertson E. J. (1998). Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell 92, 797–808. [DOI] [PubMed] [Google Scholar]
- Wicklow E., Blij S., Frum T., Hirate Y., Lang R. A., Sasaki H. and Ralston A. (2014). HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst. PLoS Genetics 10, e1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier G., Blessing M., Labosky P. A. and Hogan B. L. (1995). Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev 9, 2105–2116. [DOI] [PubMed] [Google Scholar]
- Xu J., Liu H., Lan Y., Adam M., Clouthier D. E., Potter S. and Jiang R. (2019). Hedgehog signaling patterns the oralaboral axis of the mandibular arch. eLife 8, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Beppu H., Takaoka K., Meno C., Li E., Miyazono K. and Hamada H. (2009). Antagonism between Smad1 and Smad2 signaling determines the site of distal visceral endoderm formation in the mouse embryo. Journal of Cell Biology 184, 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F. and Rossant J. (2010). FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137, 715–724. [DOI] [PubMed] [Google Scholar]
- Yang X., Li C., Xu X. and Deng C. (1998). The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proceedings of the National Academy of Sciences of the United States of America 95, 3667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Li C., Herrera P. L. and Deng C. X. (2002). Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32, 80–81. [DOI] [PubMed] [Google Scholar]
- Yuan G., Yang G., Zheng Y., Zhu X., Chen Z., Zhang Z. and Chen Y. (2015). The non-canonical BMP and Wnt/β-catenin signaling pathways orchestrate early tooth development. Development (Cambridge) 142, 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Li X., He M., Ye D., Xiong F., Amin G., Zhu Z. and Sun Y. (2020). Efficient generation of zebrafish maternal-zygotic mutants through transplantation of ectopically induced and Cas9/gRNA targeted primordial germ cells. Journal of Genetics and Genomics 47, 37–47. [DOI] [PubMed] [Google Scholar]
- Zinski J., Tajer B. and Mullins M. C. (2018). TGF-β Family Signaling in Early Vertebrate Development. Cold Spring Harb Perspect Biol 10, a033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.