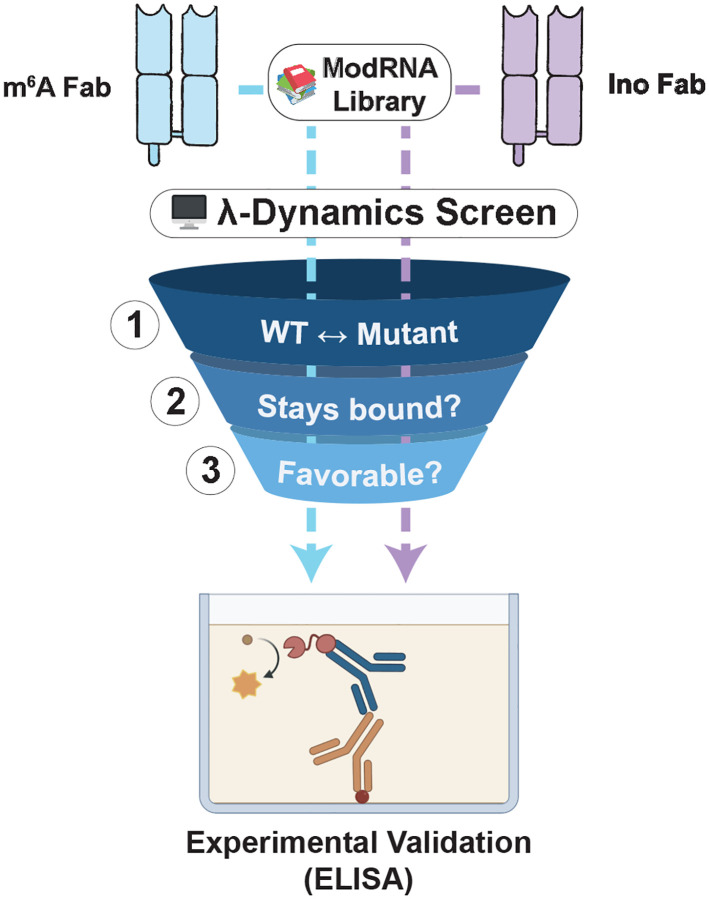
Fig 2.
In silico λ-dynamics workflow for screening potential binders to the inosine and m6A antibodies. A three-step process was used to filter candidates from a library of 48 ribonucleosides for in vitro antibody binding validation. (1) For each mutant library candidate, a λ-dynamics simulation was conducted to calculate a relative binding free energy between the mutant and its respective native ribonucleoside ligand (inosine or m6A). (2) All ribonucleosides that unbound during these simulations were deemed unfavorable and excluded from further processing. (3) Mutant bases with relative binding free energies deemed favorable (ΔΔGbind ≤ −0.7 kcal/mol) were selected for in vitro validation with binding assays based on commercial availability.