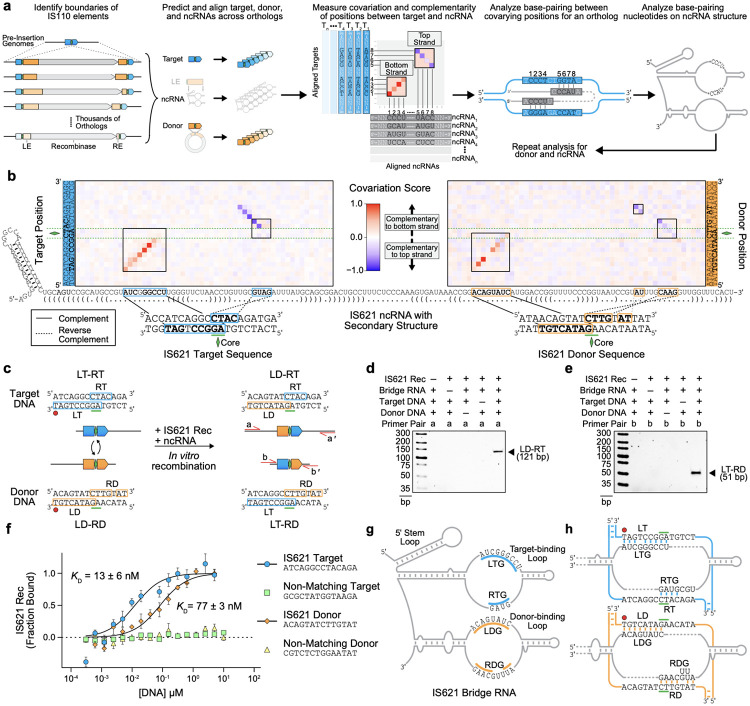
Figure 2: Computational identification of IS621 bridge RNA binding loops with sequence-specific recognition of target and donor DNA.
(a) Schematic of our computational approach to assess base-pairing potential between the IS110 ncRNA and its cognate genomic target site or donor sequence. Boundaries of thousands of IS110s were used to predict 50-bp target and donor sequences flanking the dinucleotide core (left). A quantification of covariation and base-pairing potential between target-ncRNA or donor-ncRNA pairs yielded a matrix in which predicted base-pairing interactions are depicted by diagonal stretches of red signal (indicating ncRNA complementarity to the bottom strand of the DNA) or blue signal (indicated ncRNA complementarity to the top strand of the DNA) (middle). Consensus regions of covariation were then examined for each IS110 ortholog of interest and mapped onto the predicted ncRNA secondary structure (right).
(b) Nucleotide covariation and base-pairing potential between the ncRNA and the target (left) and donor (right) sequences across 2,201 ncRNA-target pairs and 5,511 ncRNA-donor pairs, respectively. The canonical IS621 ncRNA sequence is shown across the x-axis, along with dot-bracket notation predictions of the secondary structure. Covariation scores calculated from thousands of IS110 orthologs are colored according to strand complementarity, with −1 (blue) representing high covariation and a bias toward top strand base-pairing, 1 (red) representing high covariation and a bias toward bottom strand base-pairing, and 0 indicating no detectable covariation. Regions of notable covariation signal indicating base-pairing for IS621 are boxed. Blue and orange boxes in the IS621 ncRNA sequence highlight stretches of high base-pairing potential to its native E. coli target and donor DNA, respectively. Mapping to the corresponding complementary (solid lines) or reverse complementary (dotted lines) sequences within the double-stranded target DNA (bottom left) or donor DNA (bottom right) is indicated.
(c) Schematic of in vitro recombination (IVR) reaction with IS621. Successful recombination centered at the core dinucleotide (green diamond) would result in two expected products by primer pairs and , respectively. LD, left donor; RD, right donor; LT, left target; RT, right target.
(d) Gel electrophoresis of IVR LD-RT PCR product. Results are representative of three technical replicates. Rec, recombinase.
(e) Gel electrophoresis of IVR LT-RD PCR product. Results are representative of three technical replicates. Rec, recombinase.
(f) Binding of target and donor DNA sequences by IS621 RNP containing fluorescently labeled recombinase and ncRNA. Binding of RNP with cognate or scrambled target and donor DNA sequences is measured using microscale thermophoresis (MST). Mean ± SD for three technical replicates shown.
(g) Schematic of the IS621 bridge RNA. The target-binding loop contains the left target guide (LTG) and right target guide (RTG) (blue), and the donor-binding loop contains the left donor guide (LDG) and right donor guide (RDG) (orange).
(h) Base-pairing model of the IS621 bridge RNA with cognate target and donor DNA. The target-binding loop binds both strands of the target (blue), and the donor-binding loop binds both strands of the donor (orange).