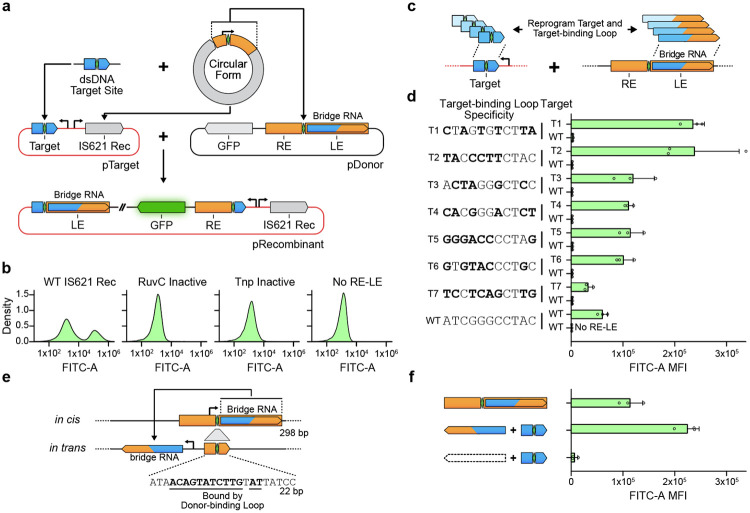
Figure 3: The IS621 target site is reprogrammable and is specified by the bridge RNA.
(a) Schematic representation of the bacterial recombination assay with bridge RNA in cis. Successful recombination centered at the core dinucleotide places GFP downstream of the synthetic promoter, resulting in fluorescence. Rec, recombinase.
(b) DNA recombination with catalytic variants of IS621 recombinase. From left to right: WT IS621; a variant of the IS621 recombinase containing the D11A/E60A/D102A/D105A mutation in the RuvC active site; a variant of the IS621 recombinase containing the S241A mutation in the Tnp domain active site; and a variant of the pDonor lacking the RE-LE. Plots are representative of 3 replicates.
(c) Schematic of reprogrammed target and bridge RNA target-binding loop sequences.
(d) Specific recombination using reprogrammed bridge RNA target-binding loop and target sequences. Target sequences are listed on the left, and the bridge RNA is reprogrammed to base-pair with the indicated sequence. Bold bases highlight differences relative to WT donor sequence. Mean ± SD for three biological replicates shown.
(e) Schematic of bridge RNA expression in trans. Top: Expression of the bridge RNA in cis from the natural promoter sequence across the RE-LE junction, as shown in (a); Bottom: Expression of the bridge RNA in trans from a synthetic promoter; transcriptional termination is achieved via a HDV ribozyme sequence. To eliminate expression of the bridge RNA in cis from the donor, the RE-LE junction in the donor is truncated to a 22 bp region lacking the −35 box of the promoter and containing the LD and RD nucleotides required for base-pairing with the bridge RNA donor-binding loop (bold and underlined).
(f) Comparison of recombination efficiency with bridge RNA expressed in cis and in trans. Mean ± SD for three biological replicates shown.