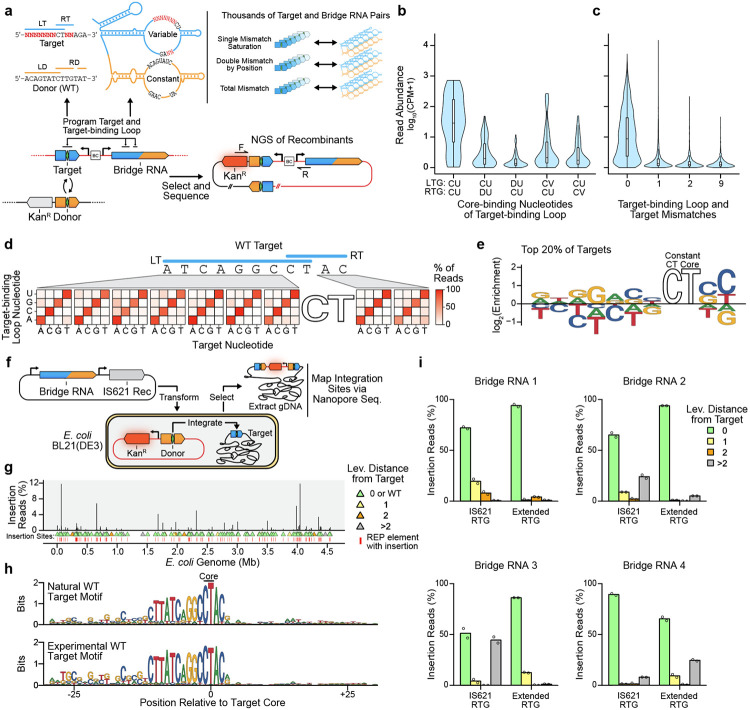
Figure 4: High-throughput characterization of IS621 target specificity demonstrates flexible programmability.
(a) Schematic representation of a target specificity screen. Successful recombination enables survival of E. coli through expression of kanamycin resistance cassette (KanR). The target sequence and bridge RNA are separated by a DNA barcode of 12 nt (BC). A T7-inducible IS621 recombinase is expressed from the plasmid bearing the target and bridge RNA. The WT donor and WT donor-binding loop are used.
(b) Mismatch tolerance between the target core and target-binding loop. Core-binding nucleotides of the target-binding loop are summarized by IUPAC codes, including D (not C) and V( not U). CPM, counts per million.
(c) Mismatch tolerance between non-core sequences of the target and target-binding loop. CPM, counts per million.
(d) Mismatch tolerance between target and target-binding loop by position. The percentage of total detected recombinants encoding each mismatch is shown. The core of the target and core-binding sequences of the target-binding loop are constant across displayed recombinants.
(e) Nucleotide enrichment of efficient perfectly matched pairs of targets and target-binding loops. The sequence logo depicts the nucleotide enrichment at each position among the top 20% most efficient targets tested in the target specificity screen.
(f) Schematic of genome insertion assay in E. coli. An E. coli cell line harbors a replication-incompetent plasmid encoding the WT donor (see Fig. 3e). Integrants are enriched by selecting for expression of kanamycin resistance from the genome after transformation with a plasmid encoding the recombinase and bridge RNA. Integration sites are determined via nanopore sequencing of isolated genomic DNA.
(g) Genome-wide mapping of insertions mediated by the WT IS621 bridge RNA. The percentage of total reads mapped to each insertion site is depicted. Triangles indicate the number of differences from the intended target site as measured by Levenshtein distance. Insertions into REP elements are highlighted with a red line.
(h) Insertion sequence preference of IS621. Sequence logos depict the target site motifs among natural (top, Methods) and experimentally observed (bottom, see Fig. 4g) IS621 target sites.
(i) Genomic specificity profile of four reprogrammed bridge RNAs. Using the same approach in (f), 11 bp target sequences appearing once in the E. coli genome were targeted. The RTG of the bridge RNA was programmed to target the RT of the 11 bp sequence (IS621 RTG, length 4 nt) or the same RT and the 3 adjacent bases (Extended RTG, length 7 nt). Color indicates the number of differences from the intended sites as measured by Levenshtein distance. Data represent sums of all insertion sites with 0, 1, 2 or >2 differences from the intended target.