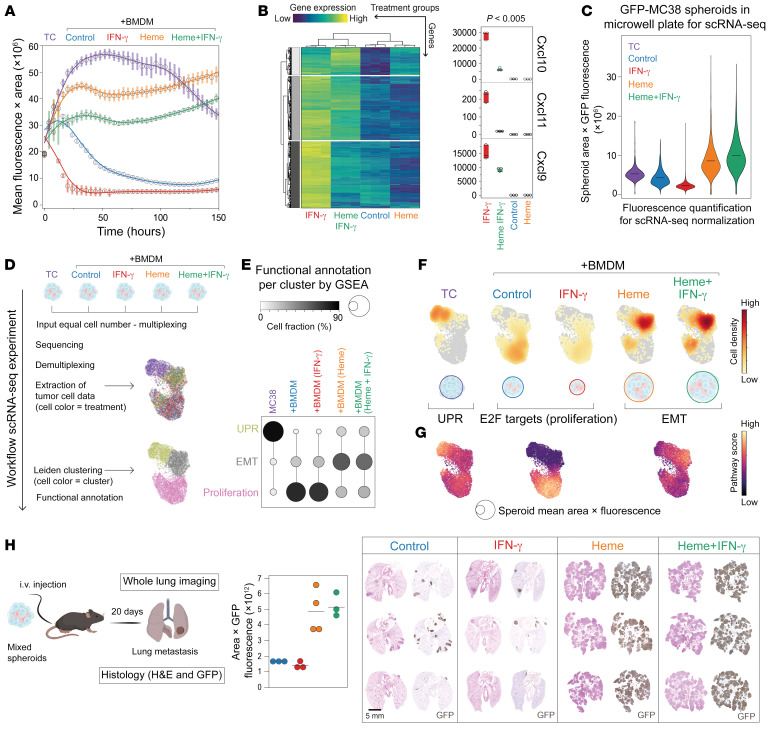
Figure 6. Heme-TAMs resist tumoricidal transformation by IFN-γ.
(A) Integrated spheroid GFP fluorescence obtained by live-cell microscopy. Data are mean ± 95% CI of 10 replicates from 1 representative experiment. (B) Expression heatmap and clustering of differentially expressed genes (log2[fold change] 0.5, P 0.005, n = 3). Right: Normalized count data for Cxcl9, Cxcl10, and Cxcl11. Each point represents 1 replicate. (C) Spheroids identical to those in A were grown in microwell plates for scRNA-Seq and scanned with a fluorescence microscope on day 9. Violin plots depict GFP fluorescence integrated across the object area for ≥1,400 spheroids per condition (ANOVA with Tukey-Kramer post-test corrected with P 0.001 for each comparison). (D) scRNA-Seq workflow. (E) GSEA-defined functional attributes for the 3 tumor cell clusters. Dot plot visualizes the fraction of tumor cells within each functional state. (F) Tumor cell densities normalized by the mean integrated fluorescence of the input. Bubbles beneath the UMAPs depict the mean spheroid size. (G) Gene expression score intensities for GSEA categories. (H) Approximately 750 spheroids (GFP-MC38 cancer cells + BMDMs) were collected from microwell plates on day 4 after spheroid formation and injected i.v. into Rag2−/−γc−/− mice. Lungs were collected 20 days after injection. Paraffin sections of lung tissue visualize metastatic disease. Scale bar: 5 mm. GFP fluorescence of whole-lung fluorescence images (see Supplemental Figure 4E) was integrated across the imaged lung area and quantified for n = 3–4 animals per condition. Each dot represents 1 mouse (lung). ANOVA with Tukey-Kramer post-test corrected for multiple comparisons, IFN-γ vs. heme + IFN-γ P = 0.0028, heme + IFN-γ vs. control P = 0.0046, heme vs. IFN-γ P = 0.0030, heme vs. control P = 0.0051, control vs. IFN-γ P = 0.98, heme vs. heme + IFN-γ P = 0.97.