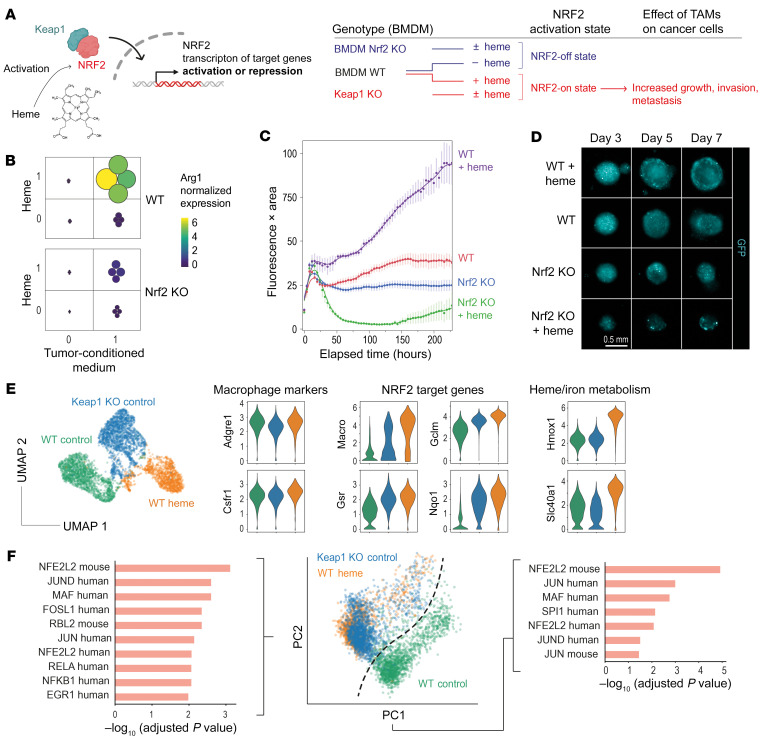
Figure 7. Heme-TAM transformation progresses via NRF2 signaling.
(A) Keap1- and Nrf2-KO mice generated BMDMs with locked NRF2-on and NRF2-off states, independent of heme exposure. (B) Factorial experiment defining the effect of heme treatment and MC38 cell culture supernatant on the expression of Arg1 mRNA measured by RT-qPCR in Nrf2-WT and Nrf2-KO BMDMs. Color and size of the dots indicate the normalized gene expression per sample (n = 4 per condition). The data demonstrate that NRF2 is required to leverage the synergistic effect of heme and tumor cell supernatant. (C) Live-cell microscopy analysis of spheroids of GFP-MC38 cancer cells mixed with Nrf2-KO and Nrf2-WT BMDMs that were untreated or pretreated with heme. Data represent the GFP fluorescence intensity integrated across the object area. Data are mean ± 95% CI of 15 replicates analyzed within 1 representative experiment. (D) Representative fluorescence images of spheroids. Scale bar: 0.5 mm. (E) Multiplexed scRNA-Seq experiment of untreated 2D cultured WT BMDMs, heme-treated WT BMDMs, and untreated Keap1-KO BMDMs. The UMAP visualizes that the interaction of genotype and treatment defines distinct gene expression patterns. The violin plots visualize the expression of canonical myeloid marker genes, NRF2-regulated genes, and heme metabolism genes as log10 (normalized count + 1) values. (F) PCA of the transcriptome data described in E. The genes defining PC1 and PC2 were analyzed for driver transcription factors by EnrichR using the TRRUST Transcription Factors 2019 data set. This analysis indicates that heme-treated BMDMs and Keap1-KO macrophages share activated NRF2 as a driver of their phenotype.