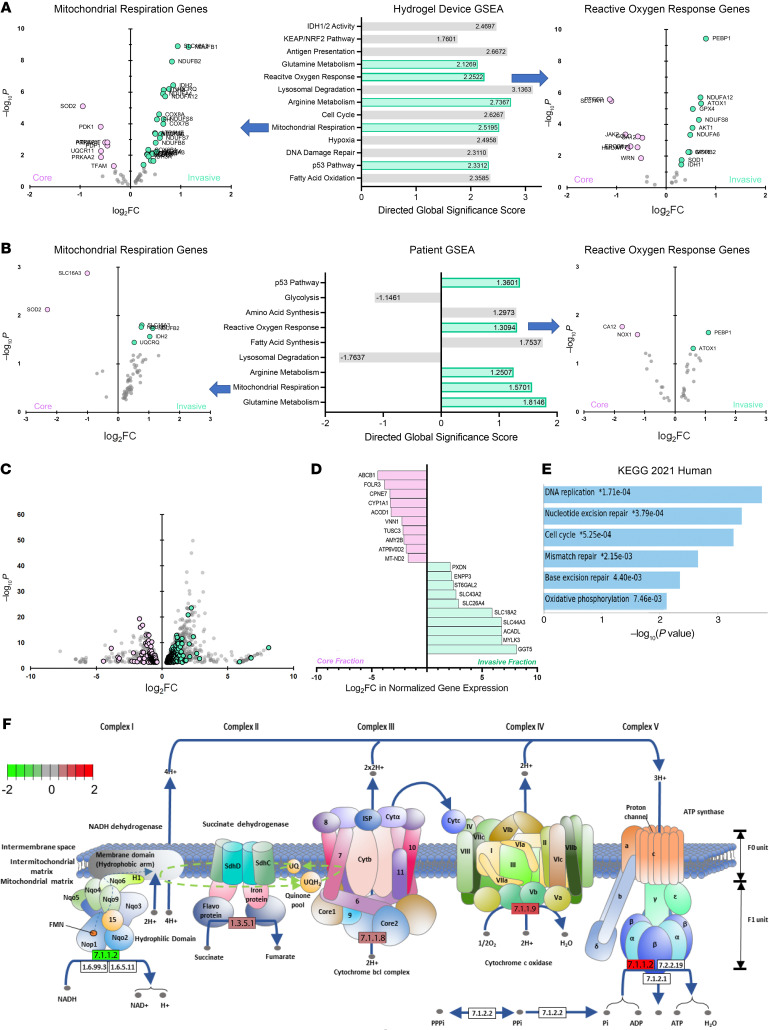
Figure 3. Gene-expression profiling demonstrates upregulated pathways involved in adapting to oxidative stress in invasive GBM cells.
(A and B) RNA from invasive and core (A) GBM43 cells from hydrogel invasion devices or (B) site-directed biopsies of patient GBMs were assessed using the NanoString nCounter panel, which analyzes expression of 770 genes from 34 metabolic pathways, with GSEA revealing enriched metabolic pathways, including 5 shared between GBM43 cells in hydrogels and patient specimens (green). Volcano plots (P and FC = probability of significance and fold change invasive versus core) are shown for genes in 2 of these pathways — mitochondrial respiration (left) and ROS response genes (right) — highlighting genes in invasive (log2FC > 0) and core (log2FC < 0) samples. (C–F) Bulk RNA-Seq on invasive and core GBM43 cells isolated from hydrogels revealed the following: (C) Of 2,172 up- or downregulated (Padjusted < 0.05) genes in invasive versus core GBM43 cells (gray dots on volcano plot), 344 (16%) were involved in cellular metabolism (green dots = upregulated genes, pink dots = downregulated genes). (D) Among 2,172 up- or downregulated (Padjusted < 0.05) genes in invasive versus core GBM43 cells (gray dots on volcano plot), shown are the 10 most up- and downregulated metabolic genes (green dots = upregulated genes, pink dots = downregulated genes and listed accordingly in the graph to the right). (E) KEGG pathway analysis of genes enriched in invasive GBM cells implicated pathways involved in the production of and response to ROS. (F) Gene-expression changes overlaid on an oxidative phosphorylation schematic revealed upregulated genes encoding mitochondrial complexes II–V in invasive GBM43 cells versus those in the core.