FIGURE 4.
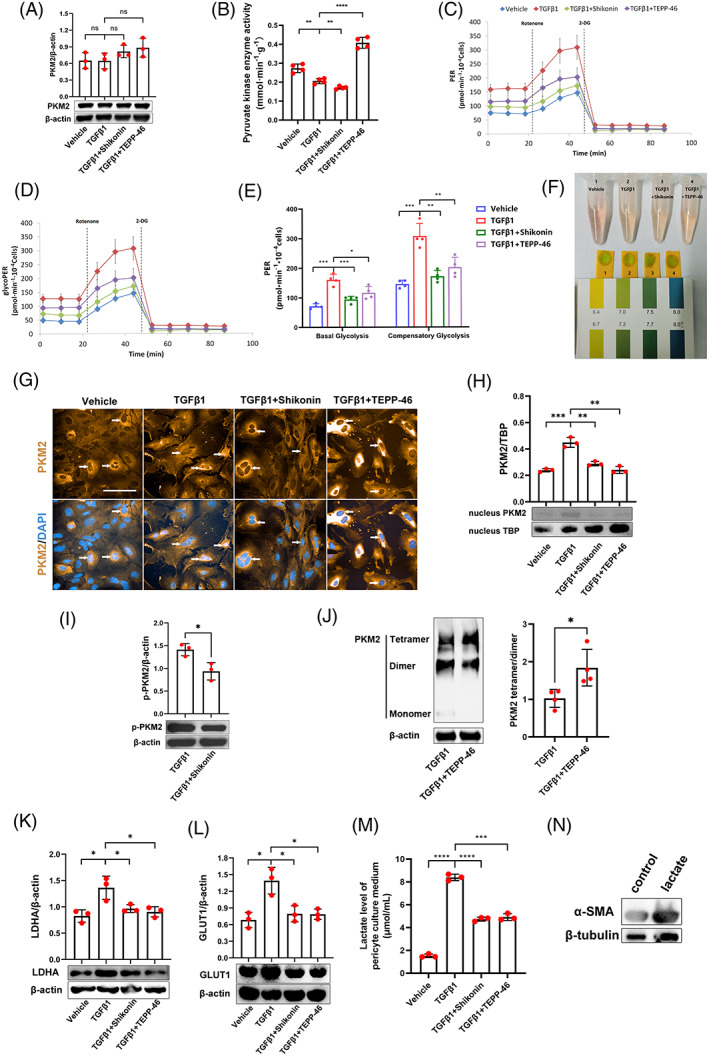
PKM2 influences renal pericyte‐myofibroblast transdifferentiation by transcription‐induced lactate production in vitro. (A) Expression of PKM2 in renal pericytes, as detected by Western blotting (n = 3). (B) Pyruvate kinase enzymatic activity in renal pericytes as investigated in vitro by pyruvate kinase enzyme activity assays (n = 4). (C,D) Total PER profile (C) and glycolytic PER profile (D) in renal pericytes, as detected by Seahorse experiments (n = 4). (E) Basal glycolysis PER and compensatory glycolysis PER of renal pericytes, as detected by Seahorse experiments (n = 4). (F) Photograph showing renal pericyte culture media and pH test strips. (G) The location of PKM2, as observed by immunofluorescent staining. White arrows indicate PKM2 in the nucleus of renal pericytes. (H) The expression of PKM2 in the nucleus of renal pericytes, as detected by Western blotting (n = 3). (I) The expression of p‐PKM2 Y105 in renal pericytes with or without Shikonin treatment, as detected by Western blotting (n = 3). (J) The expression of PKM2 in DSS crosslinked renal pericytes with or without TEPP‐46 treatment, as detected by Western blotting (n = 4). (K,L) The expression of LDHA (K) and GLUT1 (L) in renal pericytes, as detected by Western blotting (n = 3). (M) Lactate levels in pericyte culture media, as detected by a lactate assay kit (n = 3). (N) The expression of α‐SMA in renal pericytes with or without lactate treatment, as detected by Western blotting. The data are presented as follows: Error bars, mean ± SD; ns, not significant; *p < 0.05, **p < 0.01; ***p < 0.001, ****p < 0.0001; Scale bar = 100 μm. DAPI, 4,6‐diamidino‐2‐phenylindole; DSS, disuccinimidyl suberate; GLUT1, glucose transporter 1; glycoPER, glycolytic proton efflux rate; LDHA, lactate dehydrogenase A; PER, proton efflux rate; PKM2, pyruvate kinase M2; TBP, TATA binding protein; TGFβ1, transforming growth factor β1; α‐SMA, alpha smooth muscle actin.
