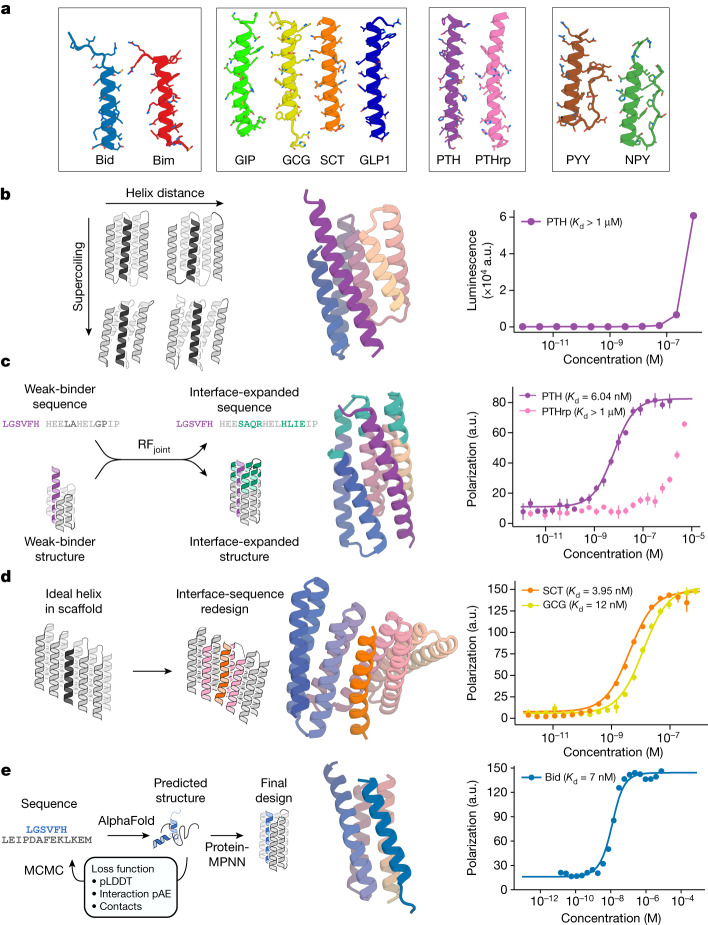
Fig. 1. Design strategies for binding helical peptides.
a, Helical peptide targets: apoptosis-related BH3 domains of Bid43 (PDB ID: 4QVE) and Bim44 (PDB ID: 3FDL), GCG3 (PDB ID: 1GCN), gastric inhibitory peptide45 (GIP; PDB ID: 2QKH), SCT46 (PDB ID: 6WZG), GCG-like peptide 147 (GLP1; PDB ID: 6X18), PTH48 (PDB ID: 1ET1), PTHrp25 (PDB ID: 7VVJ), PYY49 (PDB ID: 2DEZ) and NPY50 (PDB ID: 7X9A). b, Parametric approach. Left: sampling groove scaffolds varying supercoiling and helix distance to fit different targets. Middle: design model (spectrum) and PTH target (purple) of the best parametrically designed PTH binder. Right: split NanoBiT titration of PTH and the binder showed weak binding. a.u., arbitrary units. c, Inpainting binder optimization. Left: redesign of parametrically generated binder designs using RFjoint Inpainting to expand the binding interface and ProteinMPNN to redesign the sequences. Middle: AF2 prediction of Inpainted design (spectrum) with extended interface (teal), and PTH target (purple). Right: FP measurements (n = 4) indicate 6.04 nM binding to PTH and weak binding to off-target PTHrp. d, Threading approach to peptide binder design. Left: starting with a helix-bound scaffold, a target is threaded onto the bound helix and the interface is redesigned. Middle: AF2 prediction of design (spectrum) and SCT target (orange). Right: FP measurements (n = 4) indicate 3.95 nM binding to SCT and 12 nM binding to GCG. e, Hallucinating peptide binders. Left: Markov chain Monte Carlo (MCMC) steps are carried out in sequence space. At each step, the peptide sequence is re-predicted, and changes are accepted or rejected on the basis of interfacial contacts and AF2 metrics. The final structure is then redesigned using ProteinMPNN to avoid adversarial sequences. Interaction pAE, predicted alignment error across the interface; pLDDT, predicted local distance difference test. Middle: AF2 prediction of design (spectrum) and Bid target (blue). Right: FP measurements (n = 4) indicate 7 nM binding affinity to Bid.