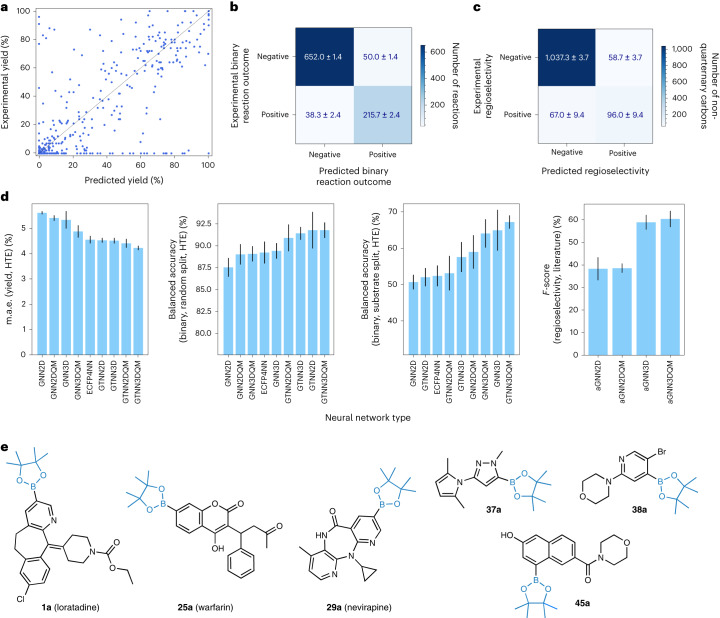
Fig. 3. Results of binary reaction outcome, reaction yield and regioselectivity predictions.
a, Performance of reaction yield prediction on the experimental dataset. The scatter plot shows predicted reaction yields on the x axis and experimental reaction yields on the y axis for GTNN3DQM. Predictions were obtained from fourfold nested cross-validation, enabling the visualization of the whole dataset (details on dataset splitting are in Supplementary Section 1). b, Confusion matrix for binary reaction outcome prediction with a threshold of ≥1% (confusion matrices with additional thresholds are in Supplementary Section 9.3). c, Confusion matrix for the prediction of non-quaternary carbons in the test set for aGNN3DQM. d, Performance of the investigated neural networks for four different tasks. Each bar plot shows the worst-performing model on the left and the best on the right. Error bars on all bar plots show the standard deviation observed on a threefold cross-validation of independent neural network training runs on the same dataset split. The centre of the error bars denotes the mean performance observed for the threefold cross-validation. The number of predicted reaction data points in the test set (n) is annotated individually. The tasks are the m.a.e. as a percent for reaction yield prediction (left; experimental dataset, n = 239); balanced accuracy (centre left; AUC) as a percent on the binary reaction outcome prediction using the random dataset split (experimental dataset, n = 239); AUC as a percent on the binary reaction outcome prediction using the substrate-based dataset split (centre right; experimental dataset, n = 239); and the performance of the four aGNNs for regioselectivity prediction measured in terms of F-score (right; literature dataset, n = 164). e, Selected examples of validated borylation opportunities as predicted by the best-performing neural network (GTNN3DQM) binary reaction outcomes of unseen substrates for three drugs (1, 25, 29) and three fragments (37, 38, 45).