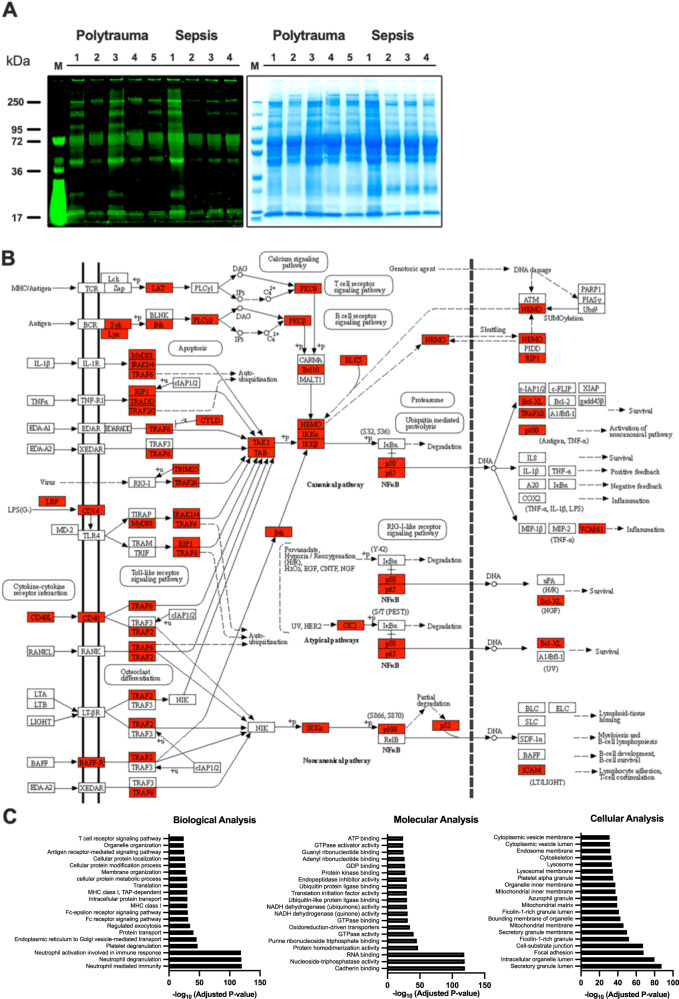
Fig. 7. Proteomic profiling and identification of differential expressed proteins in EVs isolated from polytrauma and sepsis patients.
A EV lysates were separated on SDS-PAGE, stained for phosphorylated proteins using Pro-Q Diamond staining (left panel) and total proteins were stained using Gel Code Blue Safe Protein Stain (right panel). Polytrauma (n = 5) and sepsis patients (n = 4). B NF-κB pathway involved EV identified proteins in MALDI were highlighted in red, using KEGG pathway analysis (pooled from 5 individual samples). C Gene ontology enrichment analysis of total identified proteins in MALDI-TMT analysis is shown. GO biological process, molecular process, and cellular component, enrichment analyses were performed using Enirchr web-based software, with the top 20 enriched GO terms displayed. Pooled EVs isolated from healthy individuals were used for normalization. The adjusted p value is computed using the Benjamini-Hochberg method for correction for multiple hypotheses testing.