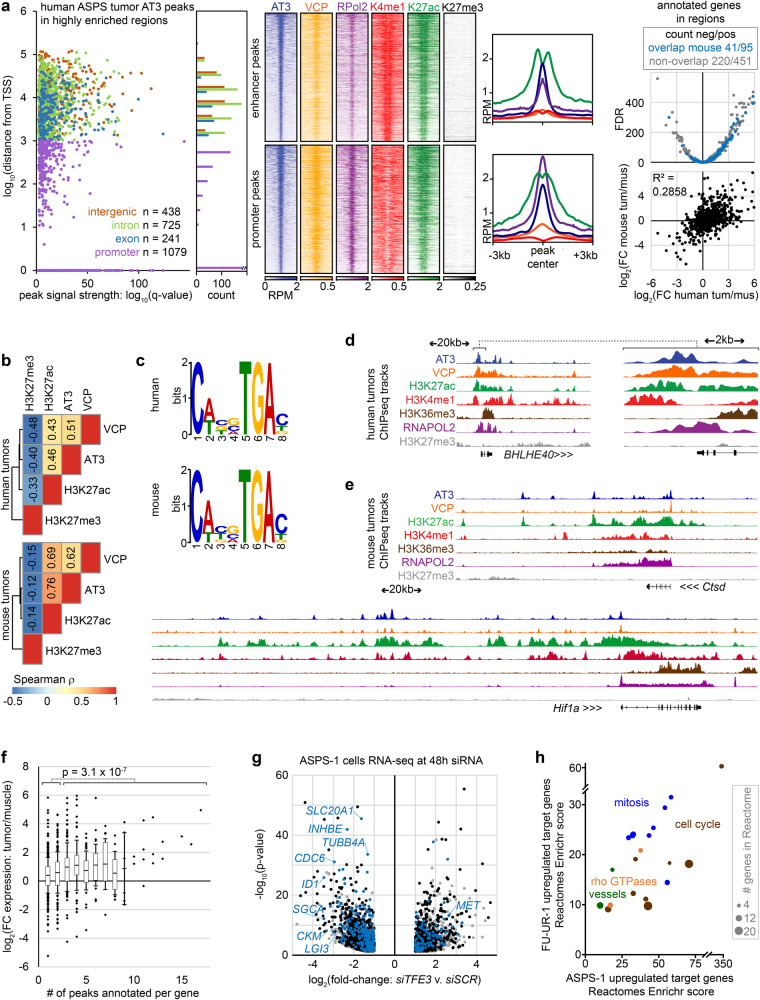
Fig. 4. AT3:VCP interaction localizes to the promoters and enhancers of target genes.
a Graph and annotation histogram for AT3 ChIP-seq peaks called in 2 human ASPS tumors within highly enriched regions (definition Supplementary Fig. 4a). RPM heatmaps for distal (putative enhancer; 1404 peaks) and proximal promoter peaks (1079 peaks; H3K4me1 window ±10 kb, others ±3 kb), and enrichment plots (H3K27me3 non-significant), then mean differential expression of nearest genes in human tumors compared to muscle samples by RNAseq (dataset GSE54729, FDR = false discovery rate q-value), noting genes also annotated in mouse highly enriched AT3 regions and correlation human to mouse differential expression tumors over muscle. b Spearman rank correlation coefficients between AT3 ChIP-seq enrichment genome-wide compared to VCP, H3K27ac and H3K27me3, (For human tumors, S = 2.81 × 1013, 3.24 × 1013, 8.45 × 1013, respectively; all p-values < 2.2 × 10−16). c Dominant motifs in peaks co-enriched for AT3, H3K27ac, and RNAPOL2. d Example ChIP-seq tracks from human tumors (n = 2, pooled) showing the region and 10-fold-magnified view (genomic distance scale noted for each). Vertical track scales are AT3: 0–4, VCP: 0–2, H3K27ac: 0–5, H3K4me1: 0–1, H3K36me3: 0–1, RNAPOL2: 0–6, H3K27me3: 0–0.2. e Example ChIP-seq tracks for Ctsd and Hif1a in mouse AT3-induced tumors. Vertical track scales AT3: 0–10, VCP: 0–4, H3K27ac: 0–9, H3K4me1: 0–2, H3K36me3: 0–2, RNAPOL2: 0–10, H3K27me3: 0–2. f Plots of mean fold-change expression in human ASPS tumors over muscle (25th to 75th percentile boxes and error bars of standard deviation, outliers depicted individually; n = 5, 3, respectively) for each number of peaks associated by nearest-gene annotations. (two-tailed heteroscedastic t-test p-value comparing 1–2 peaks to 3 or more). g Differential gene expression following 48-h siRNA depletion of AT3 in ASPS-1 cells. Blue dots represent genes annotated by highly enriched regions (from Supplementary Fig. 4a). Genes noted in black are those annotated by AT3 peaks generally. Gray dots have no associated AT3 ChIP-seq peaks. (Wald test p-values, Benjamini-Hochberg correction.) h Enrichr plot of Reactome gene sets enriched in both FU-UR-1 and ASPS-1 direct target genes of highly enriched areas and downregulated on AT3 depletion in each cell line. Specific Reactomes were assigned to general categories.