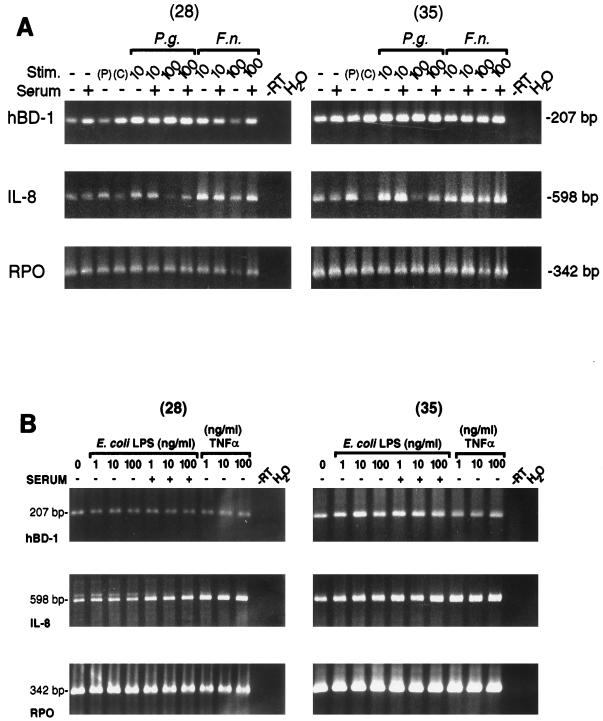
FIG. 3.
(A) Constitutive expression of hBD-1 mRNA in cultured HGE cells stimulated (Stim.) either with 10- or 100-μg/ml P. gingivalis (P.g.) or F. nucleatum (F.n.) cell wall extract in the presence or absence of 1% human serum or with 10-ng/ml PMA (P) overnight. C represents the control lane for PMA (only dimethyl sulfoxide, used as the vehicle for PMA, was added). PCR amplification was performed for 28 or 35 cycles by using primer pairs for hBD-1 (5.1-3.1 primer pair), IL-8 (a control marker indicating cellular response), and RPO, a control housekeeping gene for equivalent loading. It was evident that the apparent decrease in hBD-1 mRNA from HGE cells challenged by 100-μg/ml F. nucleatum cell wall extract in the absence of serum resulted from loading less of the RNA sample in the RT reaction mixture, since this lane gave a consistently lower signal. The sizes of PCR products were as predicted. Note the reduced signal for IL-8 with the higher dose of P. gingivalis. (B) Constitutive expression of hBD-1 mRNA in cultured HGE cells stimulated overnight either with various doses (1, 10, or 100 ng/ml) of E. coli LPS in the presence or absence of 1% human serum or with TNF-α (1, 10, or 100 ng/ml) in the absence of human serum. Three different primer pairs were included in the PCR amplification for 28 or 35 cycles, as for panel A. Note that hBD-1 expression is consistent in all lanes.