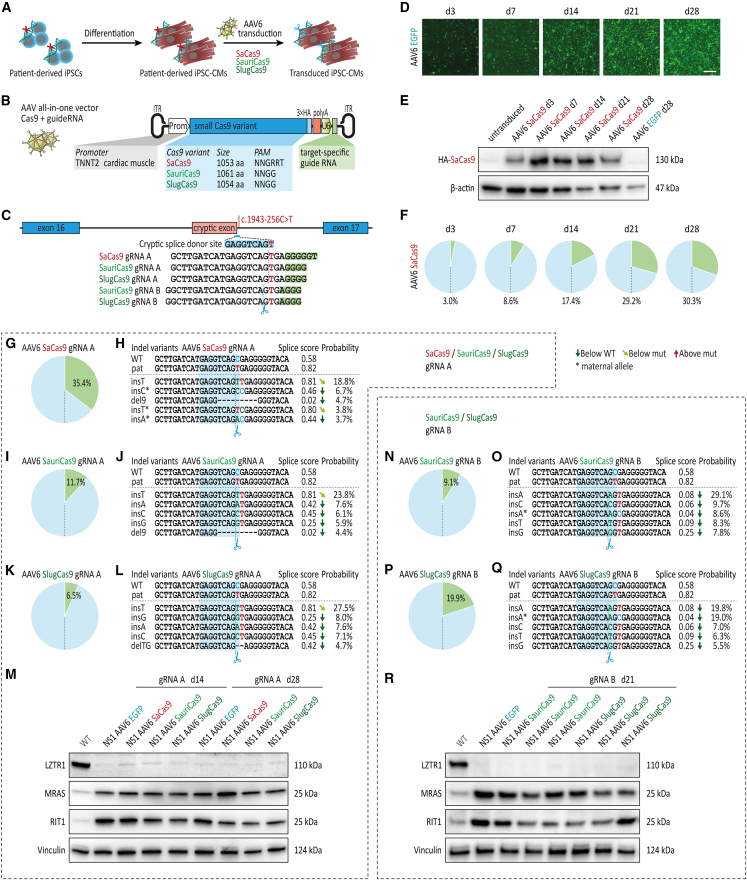
Figure 5.
All-in-one AAVs for efficient CRISPR-Cas9 editing of LZTR1 intron 16 in patient-specific iPSC-CMs
(A) Patient-specific iPSC-CMs at day 30 of differentiation were transduced with all-in-one AAVs serotype 6 encoding small Cas9 orthologs for evaluation CRISPR-Cas9 gene editing in differentiated iPSC-CMs at different time points post-transduction; n = 2–3 independent transductions per AAV construct. (B) Schematic presentation of all-in-one AAVs encoding small Cas9 orthologs under control of the cardiomyocyte-specific TNNT2 promoter together with a guide RNA expression cassette. (C) Depiction of the genome editing approach for allele-specific targeting of the deep-intronic variant in LZTR1 intron 16 in patient-specific iPSC-CMs using different small Cas9 orthologs. (D) Representative images of patient-specific iPSC-CMs transduced with AAV6-EGFP at indicated time points post-transduction; scale bar, 500 μm. (E) Representative blots of SaCas9 levels, assessed by western blot, in patient-specific iPSC-CMs transduced with AAV6-SaCas9-guide RNA A at indicated time points post-transduction; β-actin served as loading control. (F) Analysis of editing efficiency, assessed by amplicon sequencing, in patient-specific iPSC-CMs transduced with AAV6-SaCas9-guide RNA A at indicated time points post-transduction. (G) Analysis of editing efficiency, assessed by amplicon sequencing, in patient-specific iPSC-CMs transduced with AAV6-SaCas9-guide RNA A 4 weeks post-transduction. (H) Analysis of indel variant probabilities generated by SaCas9 and guide RNA A and computational prediction of splice site motifs in AAV-transduced iPSC-CMs. (I) Analysis of editing efficiency, assessed by amplicon sequencing, in patient-specific iPSC-CMs transduced with AAV6-SauriCas9-guide RNA A 4 weeks post-transduction. (J) Analysis of indel variant probabilities generated by SauriCas9 and guide RNA A and computational prediction of splice site motifs in AAV-transduced iPSC-CMs. (K) Analysis of editing efficiency, assessed by amplicon sequencing, in patient-specific iPSC-CMs transduced with AAV6-SlugCas9-guide RNA A 4 weeks post-transduction. (L) Analysis of indel variant probabilities generated by SlugCas9 and guide RNA A and computational prediction of splice site motifs in AAV-transduced iPSC-CMs. (M) Representative blots of LZTR1, MRAS, and RIT1 levels, assessed by western blot, revealed no apparent restoration of LZTR1 function in patient-specific iPSC-CMs transduced with small Cas9 orthologs with guide RNA A 2 and 4 weeks post-transduction in comparison with iPSC-CMs transduced with AAV6-EGFP; Vinculin served as loading control. (N) Analysis of editing efficiency, assessed by amplicon sequencing, in patient-specific iPSC-CMs transduced with AAV6-SauriCas9-guide RNA B 3 weeks post-transduction. (O) Analysis of indel variant probabilities generated by SauriCas9 and guide RNA B and computational prediction of splice site motifs in AAV-transduced iPSC-CMs. (P) Analysis of editing efficiency, assessed by amplicon sequencing, in patient-specific iPSC-CMs transduced with AAV6-SlugCas9-guide RNA B 3 weeks post-transduction. (Q) Analysis of indel variant probabilities generated by SlugCas9 and guide RNA B and computational prediction of splice site motifs in AAV-transduced iPSC-CMs. (R) Representative blots of LZTR1, MRAS, and RIT1 levels, assessed by western blot, revealed no apparent restoration of LZTR1 function in patient-specific iPSC-CMs transduced with small Cas9 orthologs with guide RNA B 3 weeks post-transduction.