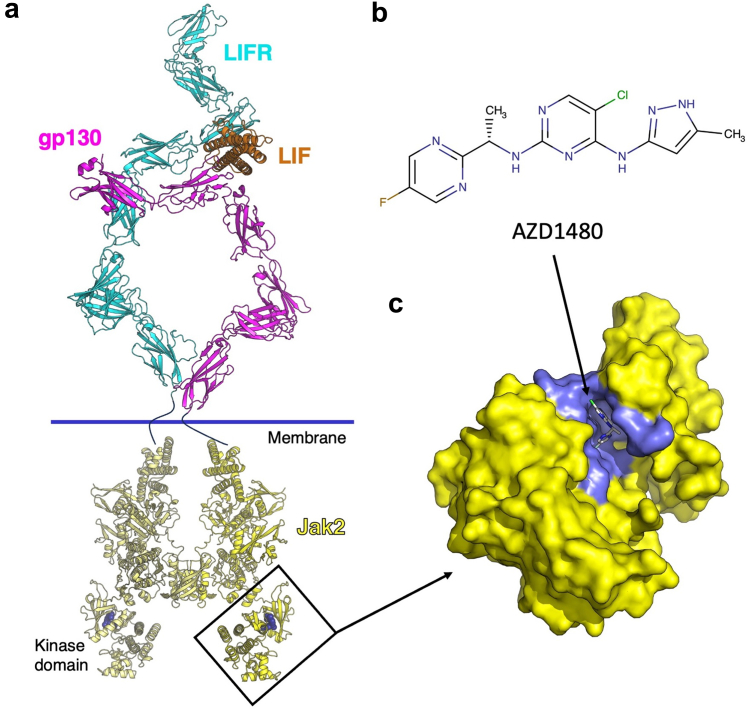
Figure 7.
LIFR, gp130, and LIF ternary complex and inhibition of the JAK/STAT signaling pathways. Upon binding to the LIFR/gp130 heterodimeric receptor complex, cytokine LIF can activate the JAK/STAT pathway, leading to the altered downstream gene expression. (a) The extracellular ternary complex of gp130/LIFR/LIF complex and a composite model of the intracellular protein JAK2 are shown in ribbon model with each protein shaded magenta (gp130), cyan (LIFR), orange (LIF), or yellow (JAK2). The inhibitor AZD1480 is shown in spheres representation (dark blue). (b) shows the Kekulé diagram of AZD1480, an inhibitor of JAK2 kinase activity used in these studies. (c) The kinase domain of JAK2 is shown in yellow surface representation with AZD-1480 bound. Amino acid residues in proximity to the inhibitor are shaded in slate color. The following protein coordinates were used to generate models in this figure: LIFR [PDB ID:2Q7N],38 and LIF/gp130 [PDB ID:1PVH].39 The initial core composite gp130/LIFR/LIF structure was generated according to methods in recent papers.19,38 Additional domains for LIFR (domains 6–8) were generated by AlphaFold,40 and domains 4–6 of gp130 were taken from PDB ID: 3L5I.41 All domains were rigidly fit to the cryo-reconstruction in (see, Zhou Y, et al.).42 To generate the homology model of JAK2, structures of JAK2 domains FERM-SH2 [PDB ID: 6E2Q],43 pseudokinase [PDB ID: 4FVQ],44 and kinase with AZD1480 [PDB ID: 2XA4]45 were aligned on the JAK1 dimeric complex [PDB ID: 7T6F].46 A composite of the JAK2 domains was submitted to SWISS-MODEL47 to fill in missing gaps between structures with the dimeric JAK1 structure as a guide. Two individual JAK2 structures were aligned to the 2 copies of JAK1 to yield the dimeric JAK2. LIF, leukemia inhibitory factor; LIFR, leukemia inhibitory factor receptor.