Abstract
Purpose
To reveal the clinical significance, pathological involvement and molecular mechanism of imprinted in Prader–Willi syndrome (IPW) in RPE anomalies that contribute to AMD.
Methods
IPW expression under pathological conditions were detected by microarrays and qPCR assays. In vitro cultured fetal RPE cells were used to study the pathogenicity induced by IPW overexpression and to analyze its upstream and downstream regulatory networks.
Results
We showed that IPW is upregulated in the macular RPE–choroid tissue of dry AMD patients and in fetal RPE cells under oxidative stress, inflammation and dedifferentiation. IPW overexpression in fetal RPE cells induced aberrant apical–basal polarization as shown by dysregulated polarized markers, disrupted tight and adherens junctions, and inhibited phagocytosis. IPW upregulation was also associated with RPE oxidative damages, as demonstrated by intracellular accumulation of reactive oxygen species, reduced cell proliferation, and accelerated cell apoptosis. Mechanically, N6-methyladenosine level of the IPW transcript regulated its stability with YTHDC1 as the reader. IPW mediated RPE features by suppressing MEG3 expression to sequester its inhibition on the AKT serine-threonine kinase (AKT)/mammalian target of rapamycin (mTOR) pathway. We also noticed that the mTOR inhibitor rapamycin suppresses the AKT/mTOR pathway to alleviate the IPW-induced RPE anomalies.
Conclusions
We revealed that IPW overexpression in RPE induces aberrant apical–basal polarization and oxidative damages, thus contributing to AMD progression. We also annotated the upstream and downstream regulatory networks of IPW in RPE. Our findings shed new light on the molecular mechanisms of RPE dysfunctions, and indicate that IPW blockers may be a promising option to treat RPE abnormalities in AMD.
Keywords: age-related macular degeneration, AMD, retinal pigment epithelium, RPE, long noncoding RNA, lncRNA, N6-methyladenosine, m6A, oxidative damages
AMD is a global leading cause of irreversible vision loss in the aged population, which severely lowers patients’ life qualities.1–3 AMD occurrence and progression are largely attributed to dysfunction and death of RPE cells in the macula, the central part of the retina responsible for sharp and detailed vision. The RPE is a monolayer of polarized, cuboidal, and pigmented cells that lies between the retina and the choroid. The RPE plays multiple roles in maintaining retinal homeostasis, including light absorption and protection against photo-oxidation; transepithelial transportation of ions, water, and nutrients; delivery, storage, and enzymatic conservation of retinoids essential for phototransduction; spatial buffering of ions and fluid in the subretinal space; phagocytosis of shed photoreceptor membranes; and secretion of various growth factors.4 As a postmitotic tissue unable to regenerate, RPE cells must fulfill these functions for the life of an individual. The proposed mechanism for RPE abnormalities in AMD posits increased intracellular oxidative stress that damages cell components,5 mitochondrial dysfunction,6 irregular lipid metabolism,7 and inflammation at RPE–choroid interface.5 Currently, the main clinical treatment for AMD is intravitreal injections of anti-VEGF drugs, which solely inhibits choroidal neovascularization in neovascular AMD without restoring RPE functions. No efficient therapeutic strategy that targets RPE anomalies in AMD has been developed. Thus, deep and extensive investigations on molecular events initiating RPE degeneration are warranted to come up with novel therapeutic approaches for AMD.
Long noncoding RNAs (lncRNAs) represent an increasingly studied class of RNA molecules that are longer than 200 nucleotides, but have little or no protein-coding potentials.8 LncRNAs demonstrate extensive biological activities in retina, including crucial roles in retinal development, homeostasis, and function.9 Dysregulations or sequence variations of lncRNAs contribute to various retinal disorders including AMD9,10; thus, the field of noncoding RNA therapy is emerging as a next-generation approach to target diverse retinopathies.11–13 Imprinted in Prader–Willi syndrome (IPW) is a paternally expressed lncRNA located on chromosome 15q11.2, within the critical region of the Prader–Willi syndrome (PWS) locus.14 IPW is highly conserved among primates, but shows little conservation between humans and mice.14 Aberrant IPW expression contributes to diseases including PWS,14 distinct types of cancers,15,16 and choroidal neovascularization progression.17 However, the pathogenic involvement of IPW in RPE anomalies is poorly understood.
In this study, we revealed the roles of IPW in inducing AMD progression as well as triggering aberrant apical–basal polarization and oxidative damages in RPE. Mechanically, YTH N6-methyladenosine RNA binding protein C1 (YTHDC1) directly bounded to N6-methyladenosine (m6A) sites on IPW to regulate its stability. IPW interrupted RPE features by suppressing the expression of the maternally expressed 3 (MEG3) gene to sequester its inhibition on the AKT serine-threonine kinase (AKT)/mammalian target of rapamycin (mTOR) pathway. Blockers on IPW may be a promising option to treat RPE abnormalities in AMD.
Methods
Human Specimen and Ethical Approval
Human eyes of aborted fetuses (11–12 weeks gestation) were collected at The First Affiliated Hospital of Nanjing Medical University. All procedures were conducted in accordance with standard protocols of eye donation for research and were approved by the institutional ethics committees (Approval No.: 2017-SR-253.A2 by The First Affiliated Hospital of Nanjing Medical University). The study followed the Declaration of Helsinki. Written informed consent was obtained from all donors before donation.
Culture and Manipulations of Fetal RPE (fRPE) Cells
fRPE cells were isolated according to a previously established protocol.18 Intact human eyes were carefully dissected from aborted fetuses with periocular tissues and anterior segments removed. Vitreous and neural retinas were separated from eye cups and discarded, and the remaining choroid was peeled from the sclera to obtain RPE-choroid explants. Small RPE sheets were isolated, collected, digested with trypsin, and planted on laminin-coated dishes. Cells were cultured in DMEM/F12 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA), SB-431542 (10 µM; MedChemExpress, Monmouth Junction, NJ, USA), 1% N1 supplement (Sigma-Aldrich), penicillin (100 U/mL; Invitrogen), streptomycin (100 U/mL; Invitrogen), 1% nonessential amino acids (Invitrogen), taurine (250 µg/mL; Sigma-Aldrich), hydrocortisone (20 ng/mL; Sigma-Aldrich), and tri-iodothyronine (13 ng/L; Sigma-Aldrich). In the following content, complete medium was used short for supplemented culture medium.
To perform sodium iodate (NaIO3) and TNF-α assays, fRPE cells were treated with NaIO3 (250 µg/mL; Sigma-Aldrich) for 96 hours and TNF-α (100 ng/mL; Sigma-Aldrich) for 48 hours, respectively. The medium was changed every 24 hours with the addition of NaIO3 or TNF-α. For actinomycin D assay, cells treated with actinomycin D (2 µg/mL; Sigma-Aldrich) were collected at 0, 2 and 4 hours after treatment. For rapamycin treatment, fRPE cells cultured in complete medium supplemented with rapamycin (10 µM; MedChemExpress) were harvested at 48 and 72 hours after treatment for RNA and protein extraction, respectively. To conduct transfection assay, fRPE cells were seeded into six-well plates (Catalog No.: 3736; Corning, Corning, NY, USA), transfected with small interfering RNA (siRNA) (100 pmol; RiboBio, Guangzhou, China) at 50% to 60% confluence using Lipofectamine 3000 transfection reagent (Invitrogen), and were collected at 48 hours after transfection for RNA extraction.
In Vitro Differentiation of Human-Induced Pluripotent Stem Cells (hiPSCs)
In vitro differentiation of hiPSC into RPE cells was conducted per the previously described SFEB/CS method.19 The hiPSC (IMR90-57, a kind gift from Prof Guoping Fan, ShanghaiTech University, Shanghai, China) were grown on mouse embryonic fibroblasts (Innovative Cellular Therapeutics, Shanghai, China) in six-well plates (Catalog No.: 3736; Corning). Cells were maintained in DMEM/F12 medium (Sigma-Aldrich) supplemented with knockout serum replacement (20%; Gibco, Carlsbad, CA, USA), β-mercaptoethanol (0.1 mmol/L; Sigma-Aldrich), L-glutamine (2 mmol/L; Gibco), nonessential amino acids (0.1 mmol/L; Gibco), and zebrafish basic fibroblast growth factor (100 ng/mL; R&D Systems, Minneapolis, MN, USA). Two small molecule compounds CKI-7 (5 µM) and SB-431542 (5 µM), perfect substitutions for recombinant Dkk1 and Lefty-A proteins, were added during suspension culture to block the Wnt and Nodal signaling pathways.
Reagents and siRNAs
Empty lentiviral plasmid (Plv4ltr-Puro-CMV-NC; L-EV) and lentiviral plasmid containing the human IPW gene (Plv4ltr-Puro-CMV-IPW; L-IPW) were provided by KerryBio (Nanjing, China). Lentiviral plasmid L-EV or L-IPW was transfected into fRPE cells using Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer's protocol. Plasmids of the dCas13b-ALKBH5, dCas13b-ALKBH5H204A, guide RNA (gRNA), and nontargeting gRNA (NT-gRNA) were kind gifts from Prof Hong-Sheng Wang (Sun Yat-sen University). Detailed information of gRNAs was listed in Supplementary Table S1. Sequences of all gRNAs were uploaded to MEGABLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to avoid mismatch to unexpected mRNAs in the human genome. Scramble siRNA and YTHDC1-siRNA were purchased from RiboBio with sequences detailed in Supplementary Table S1.
FISH
Cy3-labeled RNA of U6, 18S rRNA, and L-IPW probe mixes for FISH were constructed by RiboBio. Subcellular location and expression of RNA transcripts were determined using the FISH kit (RiboBio) according to the manufacturer's protocol. Collected cells were fixed in 4% paraformaldehyde (PFA; Sigma-Aldrich), permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) on ice, and subsequently hybridized with Cy3-labeled FISH probe mixes. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Images were taken using an LSM 510 confocal microscope (Carl Zeiss, Jena, Germany).
RNA Isolation and Sequencing
Total RNA was extracted from harvested cells using TRIzol reagent (Invitrogen). Nuclear and cytoplasmic fractions were isolated and extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Waltham, MA, USA). RNA concentration and purity were assessed using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). cDNA was generated using a PrimeScript RT Kit (Takara, Otsu, Shiga, Japan). Quantitative PCR (qPCR) was performed to determine RNA levels using FastStart Universal SYBR Green Master (ROX; Roche, Basel, Switzerland) with StepOne Plus Real-Time PCR System (Applied Biosystems, Darmstadt, Germany). Glyceraldehyde-3-phosphate dehydrogenase and U6 expression were analyzed in parallel for normalization. Primers information was detailed in Supplementary Table S2. Sanger sequencing was performed using the Sanger Sequencing Kit (Applied Biosystems) and the 3500 Series Genetic Analyzers (Applied Biosystems) per the manufacturer's protocol.
Immunofluorescence Staining
For immunofluorescence staining, cells were seeded into 24-well plates (Catalog No.: 3524; Corning) at a density of 500 cells/mm2. Collected cells were washed with PBS, fixed in 4% PFA for 30 minutes, permeabilized with 0.5% Triton X-100 for 30 minutes, and blocked with 5% BSA for 30 minutes. Washed cells were sequentially incubated with primary antibodies at 4°C overnight and with corresponding fluorescence-conjugated secondary antibodies (dilution: 1:1000; Invitrogen) at room temperature for 2 hours. Information of primary antibodies is detailed in Supplementary Table S3. Cell nuclei were counterstained with DAPI (Sigma-Aldrich). Fluorescence was observed using an LSM5 Live confocal microscope (Carl Zeiss) and quantified with Image J software.
RNA Transcriptome Sequencing and Data Analyses
For RNA transcriptome sequencing (RNA-seq), RNA was extracted with its concentration and purity measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), and its integrity was assessed using a RNA Nano 6000 Assay Kit on the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). A total amount of 1 µg RNA was sent for sequencing library generation using the Hieff NGS Ultima Dual-mode mRNA Library Prep Kit for Illumina (Yeasen Biotechnology, Shanghai, China) following the manufacturer's recommendations. The library fragments were then purified with the AMPure XP system (Beckman Coulter, Brea, CA, USA), and sequenced on an Illumina NovaSeq platform to generate 150-bp paired-end reads, according to the manufacturer's instructions. The raw data were subsequently processed with the BMKCloud online bioinformatic platform (www.biocloud.net) for quality control, reads mapping, estimation of gene expression, and differential expression analysis. Gene functional annotations based on distinct databases were further conducted, including Gene Ontology (GO) enrichment analysis (http://geneontology.org/), Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis (https://www.genome.jp/kegg/), and gene set enrichment analysis (https://www.gsea-msigdb.org/gsea/index.jsp).
Protein Extraction and Immunoblotting
For immunoblotting, cells grown on 6-well plates (Catalog No.: 3736; Corning) were harvested and washed on ice with PBS. A total of 120 µL lysis buffer (Beyotime, Shanghai, China) supplemented with protease and phosphatase inhibitor cocktail (Beyotime) was added into each well for cell lysis. The lysate was then centrifuged at 14,000 rpm at 4°C for 25 minutes to separate the soluble proteins. The supernatant was collected with the protein concentration measured using a bicinchoninic acid assay kit (Beyotime) according to the manufacturer's instructions. The absorbance at 595 nm was determined using a Multiskan FC spectrophotometric plate reader (Thermo Fisher Scientific).
Isolated proteins were further segregated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto polyvinylidene fluoride membranes (Millipore, Boston, MA, USA). Membranes were blocked with 5% nonfat dry milk (for nonphosphorylated proteins) or 5% BSA (for phosphorylated proteins) in Tris-Buffered Saline and Tween 20 for 1 hour. Blocked membranes were then sequentially incubated with primary antibodies at 4°C overnight and corresponding horseradish peroxidase-conjugated secondary antibodies (dilution: 1:10000; ICL, Newberg, OR, USA) at room temperature for 2 hours. Primary and secondary antibodies were diluted in the blocking buffer. Information and dilution of primary antibodies are detailed in Supplementary Table S3. Blots were further washed with Tris-Buffered Saline and Tween 20 and developed with the Tanon-5200Multi Chemiluminescent Imaging System (Tanon Science & Technology, Shanghai, China). Proteins were quantified using Image J software. Levels of nonphosphorylated proteins were normalized to a loading control (glyceraldehyde-3-phosphate dehydrogenase), and expression of phosphorylated proteins (p-AKT/p-mTOR) was normalized to the total proteins (AKT/mTOR).
Analysis of Phagocytosis
To measure phagocytosis of fRPE cells, carboxylate-modified polystyrene latex beads (1 mm in diameter; Sigma-Aldrich) with yellow–green fluorescence (emission maximum, 515 nm; 70 beads per cell) were added into the culture medium of fRPE cells 12 hours before collection. Cells were then harvested and added with 0.2% trypan blue to quench extracellular fluorescence. Cell nuclei were counterstained with DAPI (Sigma-Aldrich). Fluorescence was observed with an LSM5 Live confocal microscope (Carl Zeiss) and quantified using Image J software.
Measurement of Reactive Oxygen Species (ROS)
DCFH-DA (Beyotime) was diluted in serum-free medium at the ratio of 1:1000 to achieve a final concentration of 10 mmol/L. Diluted DCFH-DA was added into fRPE cells, followed by incubation at 37°C for 20 minutes. Flow cytometric analysis was subsequently conducted using a Gallios flow cytometry (Beckman Coulter) to examine ROS-positive cells.
Cell Viability Analysis
To evaluate the viability of fRPE cells, the cell counting kit-8 (CCK-8; ApexBio, Boston, MA, USA) was used following the manufacturer's instructions. Cells were seeded into 96-well plates (Catalog No.: 3599; Corning) at the density of 500 cells/mm2. Cells were subsequently treated with 10% CCK-8 solution at 24, 48, 72, and 96 hours after transplantation, respectively. The absorbance at 450 nm was measured with a microplate spectrophotometer (Multiskan FC, Thermo Fisher Scientific) to determine cell viability.
EdU Assay
Proliferation of fRPE cells was assessed using an EdU staining proliferation kit (RiboBio) according to the manufacturer's protocol. Cells were seeded into 24-well plates (Catalog No.: 3524; Corning) at the density of 500 cells/mm2, incubated in EdU solution for 2 hours before collection, fixed in 4% PFA for 30 minutes, and permeabilized with 0.5% Triton X-100 for 10 minutes. Cells were then fluorescently labeled with nuclei counterstained using DAPI. Fluorescence intensity was measured using an upright microscope (DM4000 B, Leica, Wetzlar, Germany).
Cell Cycle Analyses
To analyze cell cycle progression, we used the cell cycle staining kit (Multisciences Biotech, Hangzhou, China) according to the manufacturer's instructions. A total of 10,000 cells from each sample were collected. Cells were suspended in DNA staining solution supplemented with permeabilization solution at room temperature for 30 minutes. Cell cycle was examined by a Gallios flow cytometry (Beckman Coulter) with data analyzed using the win cycle software.
Estimation of Cell Death
At 72 hours post transfection, fRPE cells were harvested to monitor their apoptotic rates according to a previously defined protocol.20 Cells were incubated with Annexin V-FITC (R&D Systems) and propidium iodide (R&D Systems) per the manufacturer's protocol. Apoptotic cells labeled with Annexin V-FITC were detected by a Gallios flow cytometry (Beckman Coulter). Data were displayed as two-color dot plot with Annexin V-FITC (x axis) vs. propidium iodide (y axis).
Methylated RNA Immunoprecipitation-qPCR (MeRIP-qPCR) and RIP-qPCR
MeRIP was conducted using a riboMeRIP m6A Transcriptome Profiling Kit (RiboBio) according to the manufacturer's protocol. Ten percent of total RNA was removed as input control. Protein A/G magnetic beads coated with m6A antibodies were added into the remaining RNA for MeRIP assay. RIP experiment was performed using a Magna RIP Kit (Millipore) following the manufacturer's instructions. Protein A/G magnetic beads coated with antibodies against IgG or YTHDC1 were used for RIP assay. Enrichment of m6A-immunopurified and protein-bound IPW transcripts was measured using qPCR and normalized to the input.
SELECT qPCR
To detect level of motif-specific m6A modification, SELECT qPCR was conducted using the Epi-SELECT m6A site identification kit (Epibiotek, Guangzhou, China) per the manufacturer's instructions. Primers used for SELECT qPCR are listed in Supplementary Table S2.
Quantification and Statistical Analysis
All measurements were performed in a blinded manner. GraphPad Prism (v 4.0; GraphPad Software, San Diego, CA, USA) was used for statistical analyses. Two-tailed Student t test was used for comparison between two groups. One-way ANOVA coupled with the Bonferroni's post hoc test was applied for comparisons among three groups. Data were presented as mean ± SD. A P value of less than 0.05 was considered of statistical significance. Replicate information for each experiment was detailed in figure legends.
Results
Characteristics and Roles of IPW in AMD
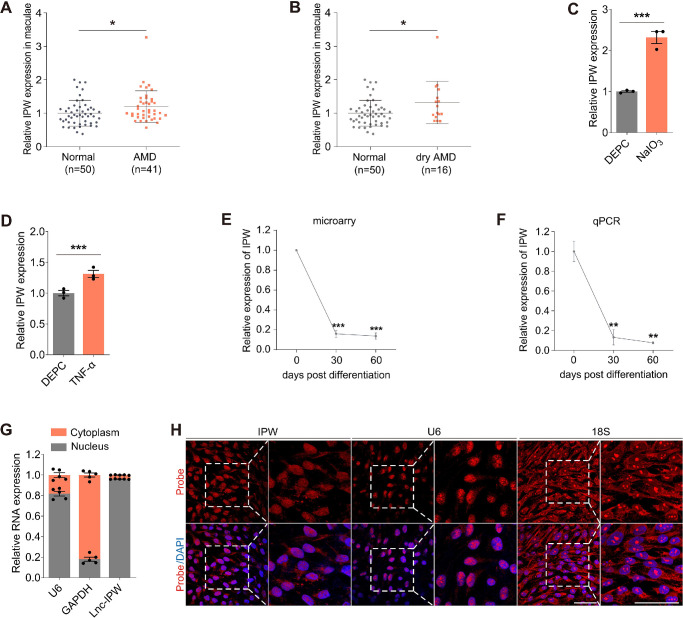
To analyze the roles of lncRNAs in AMD pathogenesis, we compared their expression levels in RPE-choroid samples of AMD patients and normal controls (data obtained from GEO database: GSE29801). Personal information and clinical diagnosis of all individuals were detailed in Supplementary Table S4. We found that expression of lncRNA IPW was upregulated in the macular RPE-choroid tissue of AMD patients compared with normal controls (Fig. 1A). Further assessment revealed that IPW expression was mainly enhanced in the macular RPE–choroid tissue of dry AMD (Fig. 1B), indicating its potential involvement in AMD, especially dry AMD.
Figure 1.
Characteristics and roles of IPW in AMD. (A, B) IPW expression detected by microarray in macular RPE-choroid complex of patients with AMD (A; n = 41) or dry AMD (B; n = 16) and age-matched controls without retinopathy (n = 50). Data was obtained from GSE29801. (C, D) IPW expression detected by qPCR in RPE cells treated with NaIO3 (C; n = 3 per group) or TNF-α (D; n = 3 per group) compared with the control group. (E, F) IPW expression detected by microarray (E; n = 3 per group) and qPCR (F; n = 3 per group) in hiPSCs and hiPSC-induced RPE cells at 30 and 60 days after differentiation. (G, H) Subcellular distribution of U6, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and IPW in fRPE cells demonstrated by qPCR (G; n = 5 per group) and FISH assay (H). Data are presented as mean ± SEM. Two-tailed Student t test was used. *P < 0.05, **P < 0.01, and ***P < 0.001.
We next measured IPW levels in RPE cells under pathogenic conditions. IPW was found upregulated in RPE cells treated with NaIO3 (Fig. 1C), an oxidative toxic agent that injures RPE in AMD,21 as well as TNF-α (Fig. 1D), a pleiotropic cytokine that promotes inflammation in AMD.22 As an initiating event in AMD, RPE dedifferentiation triggers a series of pathological changes in RPE features and functions.23 We thus analyzed IPW expression in hiPSCs and hiPSC-induced RPE cells at 30 and 60 days after differentiation, respectively. Both microarray and qPCR revealed that IPW was consistently downregulated along with RPE differentiation (Figs. 1E, 1F).
The subcellular location of a lncRNA shows significant impacts on its biological functions and molecular mechanism within the cell. Herein, qPCR revealed that IPW is predominantly expressed in RPE nucleus (Fig. 1G). Consistently, FISH assay also demonstrated endonuclear expression of IPW in RPE cells (Fig. 1H). Because lncRNAs localized to the nucleus may regulate gene expression by interacting with chromatin and transcription factors,24 our data indicated the potential role of IPW in transcriptional regulation.
IPW Overexpression in RPE Induces Aberrant Apical–Basal Polarization and Oxidative Damage
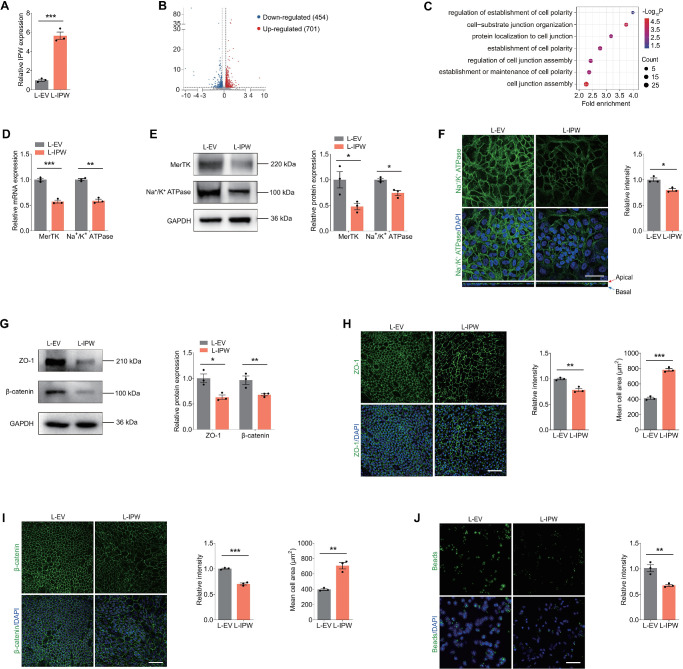
We next determined the role of IPW in regulating RPE features. We overexpressed IPW in fRPE cells by transduction of the lentivirus L-IPW. qPCR detected a 5.6-fold increase of IPW expression in fRPE cells transduced with L-IPW compared with cells transduced with L-EV (Fig. 2A). RNA-seq was also applied to explore the downstream regulatory network in fRPE cells overexpressing IPW. A total of 1155 differentially expressed genes (Log2 fold change >0.25 or <−0.25; P < 0.05) were identified, consisting of 454 upregulated and 701 downregulated genes (Fig. 2B). GO enrichment analysis based on the RNA-seq data showed dysregulation of cell polarization-related biological processes in fRPE cells transduced with L-IPW (Fig. 2C). RPE cells demonstrate a highly specialized structural and functional polarity to maintain regular functions.25 Disruption of RPE polarity contributes to progression of retinopathies including AMD.25 Herein, as revealed by qPCR and immunoblotting, both RNA and protein levels of polarized receptors MerTK and Na+/K+ ATPase were decreased in fRPE cells transfected with L-IPW (Figs. 2D, 2E). Consistently, immunofluorescence staining revealed decreased intensity of Na+/K+ ATPase in fRPE cells overexpressing IPW (Fig. 2F). The XZ image further demonstrated the change of Na+/K+ ATPase from the apical to the basolateral membrane of fRPE cells upon IPW overexpression (Fig. 2F), supporting that IPW disrupts apical–basal polarization of RPE cells.
Figure 2.
IPW overexpression in RPE induces aberrant apical–basal polarization. (A) IPW expression detected by qPCR in fRPE cells transduced with L-IPW compared with cells transduced with L-EV (n = 3 per group). (B) Volcano diagram of RNA-seq data from fRPE cells transduced with L-IPW compared with cells transduced with L-EV (log2 fold-change >0.25 or <−0.25; P < 0.05). (C) GO plot of enriched cell polarization-related biological processes in fRPE cells overexpressing IPW. (D, E) Expression of MerTK and Na+/K+ ATPase detected by qPCR (D; n = 3 per group) and immunoblotting (E; n = 3 per group) in fRPE transduced with L-EV or L-IPW. (F) Immunofluorescence staining of Na+/K+ ATPase in fRPE cells transduced with L-EV or L-IPW (n = 3 per group). Scale bar, 50 µm. (G) Immunoblotting of ZO-1 and β-catenin in fRPE cells transduced with L-EV or L-IPW (n = 3 per group). (H, I) Immunofluorescence staining of ZO-1 (H; n = 3 per group) and β-catenin (I; n = 3 per group) in fRPE cells transduced with indicated lentivirus. Scale bar, 60 µm. (J) Fluorescence of intracellular latex beads in fRPE cells transduced with indicated lentivirus (n = 3 per group). Scale bar, 60 µm. Data are presented as mean ± SEM. Two-tailed Student t test was used. *P < 0.05, ** P < 0.01, and *** P < 0.001.
We further tested whether IPW overexpression disturbs integrity of tight and adherens junctions in RPE cells, which are crucial in maintaining RPE polarity and integrity.25,26 Both immunoblotting and immunofluorescence staining revealed suppressed expression of ZO-1, which is intimately involved in the formation, organization, and regulation of tight junctions at RPE cell membrane, in fRPE cells transduced with L-IPW compared with cells transduced with L-EV (Figs. 2G, 2H). Consistently, level of β-catenin, which forms a complex with cadherin and acts as a central player in the assembly and regulation of adherens junctions, was also reduced in fRPE cells overexpressing IPW (Figs. 2G, 2I). Additionally, irregular enlargement of fRPE cells was also noticed in cells transduced with L-IPW (Figs. 2H, 2I). Intact apical–basal polarization of RPE cells is essential for maintaining proper phagocytosis of photoreceptor outer segment debris on the apical plasma membrane.25 Herein, phagocytizing ability of latex beads was inhibited in fRPE cells transduced with L-IPW (Fig. 2J), implying that IPW overexpression interrupts RPE phagocytosis. Thus, IPW overexpression in RPE cells induces aberrant apical–basal polarization typified by dysregulated polarized markers, decreased junction integrity and interrupted phagocytosis.
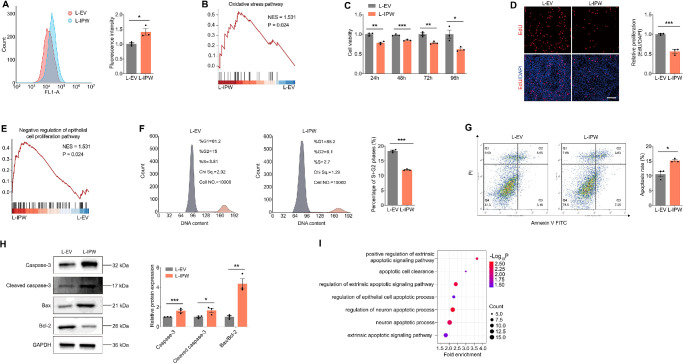
Accumulation of deleterious free radicals, induced by aberrant RPE phagocytosis, contributes to RPE oxidative damages and AMD progression.20 We, therefore, tested whether IPW overexpression in RPE cells accelerates intracellular ROS generation and induces oxidative stress injuries. ROS accumulation was detected in fRPE cells transduced with L-IPW (Fig. 3A). GSEA of the RNA-seq data identified aggravated oxidative stress in fRPE cells transduced with L-IPW (Fig. 3B). An elevated intracellular ROS level leads to oxidative-induced RPE cell death and apoptosis.27 Herein, reduced RPE cell viability was noticed in cells overexpressing IPW (Fig. 3C). EdU assay further revealed inhibited cell proliferation in RPE cells transduced with L-IPW (Fig. 3D), which was consistent with the GSEA results showing negative regulation of epithelial cell proliferation (Fig. 3E). Flow cytometric analyses demonstrated suppressed cell cycle progression in RPE cells overexpressing IPW (Fig. 3F). Flow cytometric analyses also identified the promoting effect of IPW on RPE apoptosis, as represented by increased percentage of apoptotic cells (Fig. 3G). Consistently, upregulated expression of apoptotic proteins caspase-3 and cleaved-caspase3, as well as elevated Bax/Bcl-2 ratio was detected in fRPE cells upon IPW overexpression (Fig. 3H). GO enrichment analysis based on the RNA-seq data further revealed that apoptotic processes are activated in fRPE cells transduced with L-IPW (Fig. 3I). Collectively, IPW overexpression in RPE cells leads to oxidative damages, featured by ROS accumulation as well as oxidative-induced RPE cell death and apoptosis.
Figure 3.
IPW overexpression in RPE cells induces oxidative damage. (A) ROS levels detected by flow cytometric analyses in fRPE cells transduced with L-EV or L-IPW (n = 3 per group). (B) Gene set enrichment analysis (GSEA) plot of oxidative stress pathway in fRPE cells overexpressing IPW. (C) Cell viability detected by CCK8 assay in fRPE cells transduced with indicated lentivirus (n = 3 per group). (D) Cell proliferation assessed by EdU assay in fRPE cells transduced with L-EV or L-IPW (n = 3 per group). Cell nuclei are counterstained with DAPI. Scale bar, 60 µm. (E) GSEA plot shows negative regulation of epithelial cell proliferation pathway in fRPE cells overexpressing IPW. (F) Cell cycle progression detected by flow cytometric analyses in fRPE cells transduced with indicated lentivirus (n = 3 per group). Grey area (left) indicates for G1 phase; purple area (middle) indicates for S phase; orange area (right) indicates for G2 phase. (G) Apoptosis detected by flow cytometric analyses in fRPE cells transduced with L-EV or L-IPW (n = 3 per group). (H) Immunoblotting of caspase-3, cleaved caspase-3, Bax/Bcl-2 in fRPE cells transduced with indicated lentivirus (n = 3 per group). (I) GO plot of enriched apoptosis-related pathways in fRPE cells overexpressing IPW. Data are presented as mean ± SEM. Two-tailed Student t test was used. * P < 0.05, **P < 0.01, and *** P < 0.001.
YTHDC1 Directly Binds to m6A Sites on IPW to Regulate Its Stability
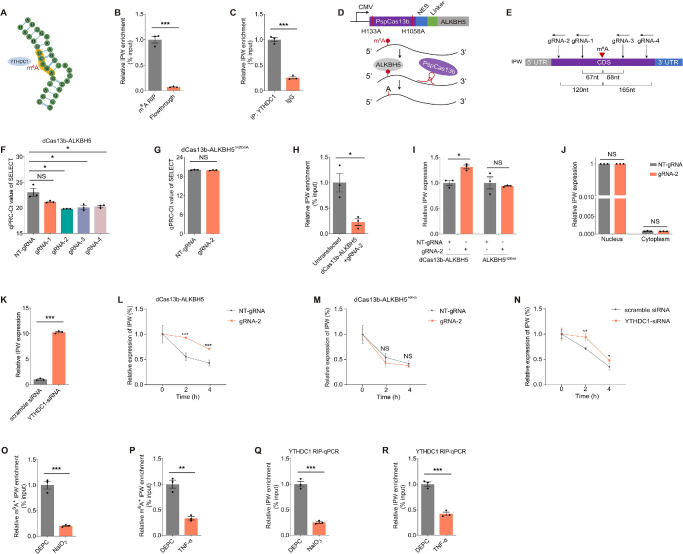
LncRNAs can be epigenetically modified by m6A modifications, which alter the structure and properties of lncRNAs, thus modulating their functions and interactions.28 We therefore annotated whether IPW is modulated by m6A modification. RMBase (https://rna.sysu.edu.cn/rmbase/index.php) and SRAMP (http://www.cuilab.cn/sramp) were applied to reveal m6A sites in IPW. An m6A site (chr15: 25119251; m6A_site_139960) shared by both online programs was identified and was predicted to bind to the endonuclear m6A reader YTHDC1 (Fig. 4A). Direct binding between the m6A antibody and m6A_site_139960 in fRPE cells was confirmed by MeRIP-qPCR experiments (Fig. 4B). Furthermore, RIP assay verified the binding between IPW and YTHDC1 (Fig. 4C), indicating that YTHDC1 potentially functions as the m6A reader.
Figure 4.
YTHDC1 directly binds to m6A sites on IPW to regulate its stability. (A) A schematic diagram demonstrating the m6A site on the IPW transcript and its interaction with YTHDC1. (B) The direct binding between the m6A antibody and m6A_site_139960 detected by MeRIP-qPCR assay in fRPE cells (n = 3 per group). (C) The binding between IPW and YTHDC1 verified by RIP-qPCR assay in fRPE cells (n = 3 per group). (D) A schematic diagram demonstrating the structure and functions of the dCas13b-ALKBH5 plasmid. (E) A schematic diagram showing specific locations of the four gRNAs (gRNA-1, gRNA-2, gRNA-3, gRNA-4) targeting m6A_site_139960 in the IPW transcript. (F) The m6A level of m6A_site_139960 detected by SELECT qPCR in fRPE cells transfected with distinct gRNAs compared with cells transfected with NT-gRNA (n = 3 per group). (G) The m6A level of m6A_site_139960 detected by SELECT qPCR in fRPE cells cotransfected with the dCas13b-ALKBH5H204A plasmid and NT-gRNA/gRNA-2 (n = 3 per group). (H) The binding between IPW and YTHDC1 detected by RIP-qPCR assay in fRPE cells cotransfected with the dCas13b-ALKBH5 plasmid and gRNA-2 (n = 3 per group). (I) IPW expression detected by qPCR in fRPE cells cotransfected with the dCas13b-ALKBH5/dCas13b-ALKBH5H204A plasmid and NT-gRNA/gRNA-2 (n = 3 per group). (J) Intracellular distribution of IPW detected by qPCR in fRPE cells cotransfected with the dCas13b-ALKBH5 plasmid and NT-gRNA/gRNA-2 (n = 3 per group). (K) IPW expression detected by qPCR in fRPE cells transfected with scramble siRNA and YTHDC1-siRNA (n = 3 per group). (L, M) Half-life of the IPW transcript detected by qPCR in fRPE cells cotransfected with dCas13b-ALKBH5 and NT-gRNA/gRNA-2 (L; n = 3 per group) and in cells cotransfected with dCas13b-ALKBH5H204A and NT-gRNA/gRNA-2 (M; n = 3 per group). (N) The stability of IPW detected by qPCR in fRPE cells transfected with scramble siRNA or YTHDC1-siRNA (n = 3 per group). (O, P) M6A level of m6A_site_139960 detected by MeRIP-qPCR in fRPE cells treated with DEPC or NaIO3 (O; n = 3 per group) or TNF-α (P; n = 3 per group). (Q, R) The binding between IPW and YTHDC1 detected by RIP-qPCR in fRPE cells treated with DEPC or NaIO3 (Q; n = 3 per group) or TNF-α (R; n = 3 per group). Data are presented as mean ± SEM. Two-tailed Student t test was used. NS, not significant (p > 0.05), * P < 0.05, ** P < 0.01, and *** P < 0.001.
To further explore the role of m6A modification in regulating IPW features, we used an engineered dCas13b-ALKBH5 fusion protein for targeted demethylation of IPW (Fig. 4D).29 Four gRNAs targeting m6A_site_139960 were designed and synthesized (Fig. 4E). We then used SELECT qPCR to verify effects of these gRNAs on IPW demethylation in fRPE cells. SELECT qPCR showed that the m6A level of m6A_site_139960 in fRPE cells is reduced by three gRNAs cotransfected with the dCas13b-ALKBH5 plasmid compared with NT-gRNA (Fig. 4F). Among all, gRNA-2 demonstrated the strongest demethylation activity and was thus selected for further assessments (Fig. 4F). However, the demethylation effect of gRNA-2 on m6A_site_139960 was abolished in fRPE cells cotransfected with the dCas13b-ALKBH5H204A plasmid carrying the catalytically inactive H204A mutation (Fig. 4G).30 RIP experiment further verified that cotransfection of the dCas13b-ALKBH5 plasmid and gRNA-2 will interrupt the binding between IPW and YTHDC1 (Fig. 4H), indicating that the direct interaction between IPW and YTHDC1 depends on the methylation status of m6A_site_139960.
Because YTHDC1 is an m6A reader protein that diversely regulates biological properties of lncRNAs,31,32 we annotated whether demethylation of m6A_site_139960 affects stability and intracellular distribution of IPW in RPE cells. Elevated IPW expression was detected in fRPE cells cotransfected with the dCas13b-ALKBH5 plasmid and gRNA-2, while such elevation was abolished in cells transfected with dCas13b-ALKBH5H204A or NT-gRNA (Fig. 4I). However, intracellular distribution of IPW was not altered by cotransfection of dCas13b-ALKBH5 and gRNA-2 (Fig. 4J), indicating that the m6A level of IPW does not affect its nuclear export. We then assessed whether YTHDC1 functions as a reader by knocking it down in fRPE cells using the previously reported siRNA.33 IPW level was found upregulated in fRPE cells transfected with YTHDC1-siRNA compared with cells transfected with scramble siRNA (Fig. 4K), suggesting that IPW expression was potentially regulated by the methylation status of m6A_site_139960 with YTHDC1 as a reader. We next analyzed whether m6A modification regulates IPW expression by modulating its RNA stability. Stability of the IPW transcript was determined using the transcription inhibitor actinomycin D. A prolonged half-life of the IPW transcript was detected in fRPE cells cotransfected with dCas13b-ALKBH5 and gRNA-2, but was not found in cells transfected with dCas13b-ALKBH5H204A or NT-gRNA (Figs. 4L, 4M). Consistent with above findings, stability of the IPW transcript was also enhanced in fRPE cells with YTHDC1 knocked down (Fig. 4N).
We further aimed to tell whether upregulation of IPW under pathogenic conditions was mediated by m6A modification. MeRIP-qPCR detected reduced m6A level of m6A_site_139960 in fRPE cells treated with NaIO3 or TNF-α (Figs. 4O, 4P). Consistently, RIP assay also revealed that the binding between IPW and the YTHDC1 protein was partly abolished in fRPE cells treated with NaIO3 or TNF-α (Figs. 4Q, 4R), which was potentially responsible for the aberrant changes in IPW expression. Collectively, our study implied that YTHDC1 directly binds to the m6A site on IPW to regulate its stability.
IPW Inhibits MEG3 to Activate the AKT/mTOR Pathway in RPE Cells
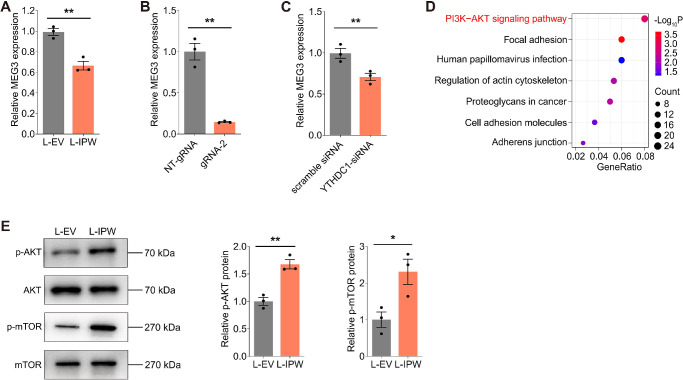
To explore the underlying mechanism of IPW in AMD, we next annotated the downstream target of IPW in RPE cells. Reportedly, IPW is a regulator of the imprinted DLK1-DIO3 locus, as it recruits the histone methyltransferase G9A to facilitate the repressive modifications of H3K9me3 at the IG-DMR of the DLK1-DIO3 region in hiPSCs.14 IPW overexpression in PWS resulted in downregulation of all maternally expressed genes (MEGs) in the DLK1-DIO3 locus, among which lncRNA MEG3 exerts a protective role against AMD by promoting RPE functions.14,34,35 Herein, we initially determined whether IPW regulates MEG3 expression in fRPE cells. Suppressed MEG3 expression was detected in fRPE cells overexpressing IPW (Fig. 5A). Moreover, MEG3 level was reduced in fRPE cells cotransfected with the dCas13b-ALKBH5 plasmid and gRNA-2 (Fig. 5B), and in cells with YTHDC1 knocked down (Fig. 5C), further indicating that IPW demethylation downregulates MEG3 expression.
Figure 5.
IPW inhibits MEG3 to activate the AKT/mTOR pathway in RPE cells. (A) MEG3 expression detected by qPCR in fRPE cells transduced with L-IPW compared with cells transduced with L-EV (n = 3 per group). (B) MEG3 expression detected by qPCR in fRPE cells cotransfected with the dCas13b-ALKBH5 plasmid and NT-gRNA/gRNA-2 (n = 3 per group). (C) MEG3 expression detected by qPCR in fRPE cells transfected with scramble siRNA or YTHDC1-siRNA (n = 3 per group). (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) plot of enriched pathways in fRPE cells transduced with L-IPW compared with cells transduced with L-EV. (E) Immunoblotting of p-AKT, AKT, p-mTOR, and mTOR in fRPE cells transduced with L-EV or L-IPW (n = 3 per group). Data are presented as mean ± SEM. Two-tailed Student t test was used. * P < 0.05 and ** P < 0.01.
We further explored the downstream regulatory network of MEG3 in RPE cells to better annotate the mechanism underlying the AMD promoting effects of the IPW-MEG3 axis. KEGG pathway enrichment assessment based on the RNA-seq data revealed the PI3K-AKT signaling pathway as the most enriched pathway in fRPE cells overexpressing IPW (Fig. 5D), which is the upstream of the AKT/mTOR signaling pathway. Although MEG3 suppresses the AKT/mTOR signaling pathway in various types of cells, including retinal endothelial cells, such regulation has not been verified in RPE cells.36–38 Activation of the AKT/mTOR pathway in RPE cells disturbs their regular functions, thus contributing to AMD onset and progression.20,23 Consistent with previous reports, we herein demonstrated that IPW overexpression activates the AKT/mTOR signaling pathway in fRPE cells, demonstrated by promoted phosphorylation of the AKT and mTOR proteins (Fig. 5E). Overall, we identified that IPW inhibits MEG3 to activate the AKT/mTOR pathway in RPE cells.
Rapamycin Suppresses the AKT/mTOR Pathway to Alleviate the IPW-induced RPE Injuries
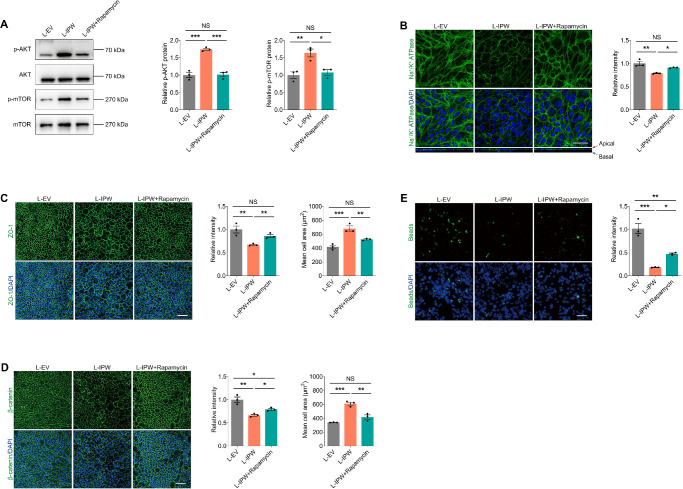
Our data imply the AKT/mTOR pathway as the downstream regulatory network of IPW in RPE cells. Rapamycin, a drug that works by inhibiting the mTOR pathway, shows protective roles in multiple retinopathies, including AMD.39,40 We herein further annotated whether IPW mediates RPE features through the AKT/mTOR pathway using rapamycin. Our data implied that rapamycin suppresses the IPW mediated activation of the AKT/mTOR pathway in fRPE cells (Fig. 6A). Immunofluorescence staining detected that the decreased fluorescence intensity and aberrant basolateral location of polarized receptor Na+/K+ ATPase induced by IPW overexpression is alleviated upon rapamycin treatment (Fig. 6B). Furthermore, the decreased expression of the tight junction marker ZO-1 (Fig. 6C) and the adherens junction marker β-catenin (Fig. 6D) in fRPE cells upon IPW transduction were restored by rapamycin. The irregular enlargement of fRPE cells was also rescued by rapamycin treatment (Figs. 6C, 6D). Additionally, the inhibited phagocytizing ability of latex beads induced by IPW in fRPE cells was partly regained after rapamycin treatment (Fig. 6E). These results indicated that the activation of AKT/mTOR pathway and aberrant apical–basal polarization of fRPE cells can be rescued by rapamycin.
Figure 6.
Rapamycin suppresses the AKT/mTOR pathway to alleviate the IPW-induced aberrant apical–basal polarization. (A) Immunoblotting of p-AKT, AKT, p-mTOR and mTOR in fRPE cells transduced with L-EV or L-IPW and treated with or without rapamycin (n = 3 per group). (B) Immunofluorescence staining of Na+/K+ ATPase in fRPE cells receiving indicated treatments (n = 3 per group). Scale bar, 50 µm. (C, D) Immunofluorescence staining of ZO-1 (C; n = 3 per group) and β-catenin (D; n = 3 per group) in fRPE cells receiving distinct treatments. Scale bar, 60 µm. (E) Fluorescence of intracellular latex beads in fRPE cells transduced with L-EV or L-IPW and treated with or without rapamycin (n = 3 per group). Scale bar, 60 µm. Data are presented as mean ± SEM. One-way ANOVA coupled with the Bonferroni's post hoc test was used. NS, not significant (P > 0.05), * P < 0.05, ** P < 0.01, and *** P < 0.001.
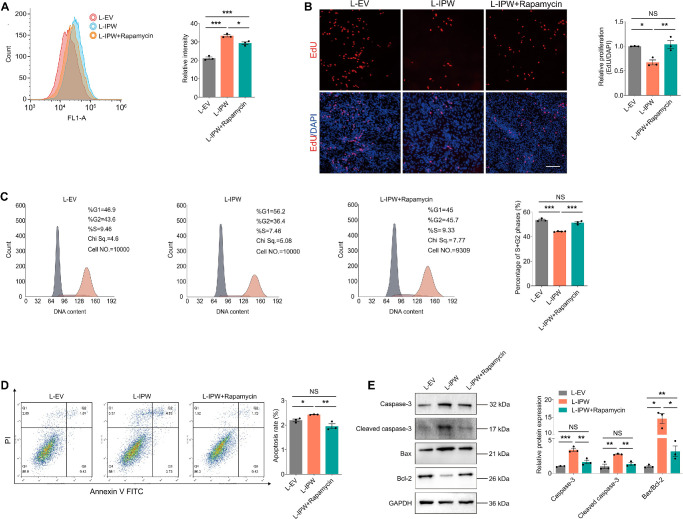
Furthermore, we tested whether rapamycin could alleviate degeneration and apoptosis in fRPE cells upon IPW overexpression. Our results showed that rapamycin also alleviated the IPW-associated ROS accumulation in fRPE cells (Fig. 7A). Inhibited proliferation of fRPE cells accompanied by suppressed cell cycle progression caused by IPW overexpression was restored by rapamycin (Figs. 7B, 7C). Moreover, IPW-induced apoptosis of fRPE cells as demonstrated by flow cytometric analyses and immunoblotting was rescued by addition of rapamycin (Figs. 7D, 7E). Collectively, the above results further implied that IPW regulates RPE features through activation of the mTOR pathway. Our data also indicated that the mTOR inhibitor, rapamycin, is a promising agent to treat IPW-induced RPE injuries.
Figure 7.
Rapamycin suppresses the AKT/mTOR pathway to alleviate the IPW-induced oxidative damage. (A) ROS accumulation detected by flow cytometric analyses in fRPE cells receiving indicated treatments (n = 3 per group). (B) Cell proliferation detected by EdU assay in fRPE cells receiving distinct treatments (n = 3 per group). Cell nuclei are counterstained with DAPI. Scale bar, 60 µm. (C) Cell cycle progression detected by flow cytometric analyses in fRPE cells transduced with L-EV or L-IPW and treated with or without rapamycin (n = 3 or 4 per group). Grey area (left) indicates for G1 phase; purple area (middle) indicates for S phase; orange area (right) indicates for G2 phase. (D) Apoptosis detected by flow cytometric analyses in fRPE cells receiving indicated treatments (n = 3 per group). (E) Immunoblotting of caspase-3, cleaved caspase-3, Bax/Bcl-2 in fRPE cells receiving distinct treatments (n = 3 per group). Data are presented as mean ± SEM. One-way ANOVA coupled with the Bonferroni's post hoc test was used. NS, not significant (P > 0.05), * P < 0.05, ** P < 0.01, *** P < 0.001.
Discussion
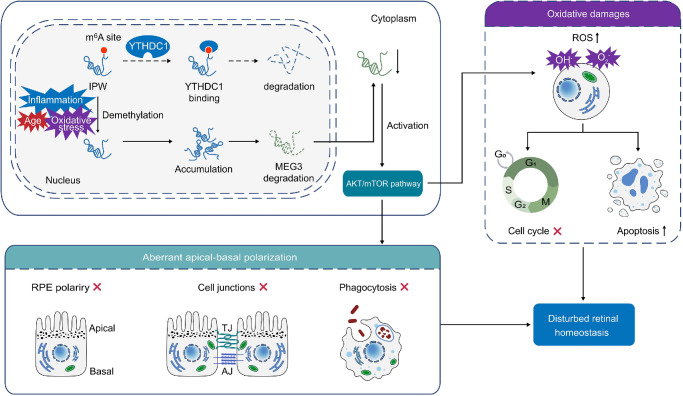
LncRNAs present crucial and extensive biological and pathological activities in the retina, and their dysregulation leads to distinct retinopathies, including AMD. Our group has previously demonstrated the involvements of lncRNAs ZNF503-AS1 and LINC00167 in RPE degeneration and AMD.41,42 In the present study, we showed that IPW is upregulated in the macular RPE-choroid tissue of dry AMD patients and in fRPE cells under pathological conditions, including oxidative stress, inflammation, and dedifferentiation. IPW overexpression in RPE caused aberrant apical–basal polarization as demonstrated by dysregulated polarized markers, disrupted tight and adherens junctions, and inhibited phagocytosis, as well as oxidative damages, as shown by intracellular ROS accumulation, decreased cell proliferation, and accelerated cell apoptosis. Mechanically, the m6A level of the IPW transcript regulated its stability with YTHDC1 as the reader. IPW mediated RPE features by suppressing MEG3 expression to sequester its inhibition on the AKT/mTOR pathway (Fig. 8).
Figure 8.
Schematic diagram of IPW-mediated effects on the RPE. Under pathogenic conditions, m6A levels in the IPW transcript are reduced, thus disturbing its degradation with YTHDC1 as a reader. IPW induces aberrant apical–basal polarization and oxidative damages in RPE cells by targeting MEG3-mediated activation of the AKT/mTOR pathway.
Aberrant apical–basal polarization and oxidative damages in RPE cells are two key factors involved in the development and progression of AMD. These pathological processes can lead to RPE dysfunction and death, impairing its ability to support and nourish the photoreceptors, ultimately leading to vision loss.43 Apical–basal polarization of RPE cells is necessary for supporting bidirectional transport of nutrients and waste products between photoreceptors and choroid, phagocytosis of photoreceptor outer segment debris, and secretion of neurotropic growth factors.44 Disruption in this polarization can disturb retinal homeostasis and health. Upon constant exposure to light and oxygen, RPE cell generate high amounts of ROS that damage cellular components such as lipids, proteins and DNA. Over time, such oxidative damages accumulate within RPE cell, further leading to its dysfunction and AMD phenotypes.33 Herein, we revealed that IPW overexpression in RPE cells contributes to both aberrant apical–basal polarization and oxidative damages, implying its role in AMD pathogenesis.
LncRNA IPW is a paternally expressed lncRNA that shows high conservation among primates. Mutations in IPW cause the neurodevelopmental disorder PWS, a rare genetic disorder that affects various aspects of a person's physical, cognitive, and behavioral development.14 Aberrant IPW expression was also detected in various types of human cancers with IPW showing both tumor promoting and suppressive impacts. IPW was downregulated in ductal carcinoma, and it inhibited ductal carcinoma growth in vitro and in vivo.15 Consistently, reduced IPW expression was identified in head-neck cancer and lung adenocarcinoma, which correlated with poor survival.16,45 Conversely, IPW was upregulated in samples of medullary thyroid cancer.46 However, the effect of IPW in AMD etiology has not been fully elucidated. Although Yang et al17 revealed that IPW silencing reduces choroidal neovascularization formation and endothelial angiogenic phenotypes, the role of IPW in regulating RPE function remains unclear. We herein detected enhanced IPW expression in the RPE-choroid samples of patients with dry AMD. We also annotated the pathological involvement of IPW in RPE anomalies, which induces aberrant apical–basal polarization and oxidative damages thus contributing to dry AMD occurrence and progression.
M6A modification, necessary for the post-transcriptional regulations of lncRNAs, plays pivotal roles in multiple biological and pathological processes.47,48 Aberrant m6A modifications of mRNAs and circular RNAs contribute to RPE degenerations and AMD pathogenesis. Reportedly, the m6A reader protein METTL3-mediated m6A modification of HMGA2 and ANXA1 mRNAs was associated with subretinal fibrosis, epithelial-mesenchymal transition, and blood–retinal barrier breakdown in neovascular AMD.49,50 The m6A eraser protein FTO alleviated Amyloid-β induced RPE degeneration via the PKA/CREB signaling pathway.51 However, effects of aberrant m6A modifications of lncRNAs on AMD occurrence and progression have not been elucidated. YTHDC1, an m6A reader protein that locates in the nucleus, has been reported to regulate various features of lncRNAs.31,32 We have previously reported that YTHDC1 mediates nuclear export of the m6A-modified circSPECC1 transcript to maintain regular functions of the RPE cells,33 although no other reports stressing the pathological involvement of YTHDC1 in AMD has been revealed. Herein, we identified another potential regulatory mechanism of YTHDC1 in RPE cells by stabilizing the IPW transcript without affecting its subcellular distributions. However, more investigations into this area are warranted.
Collectively, we identified the AMD-promoting role of IPW in RPE cells. IPW overexpression in RPE induces aberrant apical–basal polarization and oxidative damages. Mechanistically, reduced m6A levels in the IPW transcript disturbed its stability with YTHDC1 as a potential reader. IPW disturbed RPE functions through downregulation of MEG3 expression to activate the AKT/mTOR pathway. Moreover, IPW-induced RPE injuries were alleviated by the mTOR inhibitor rapamycin. Silencing of IPW is a promising therapeutic option for AMD and other retinopathies involving RPE dysfunctions.
Supplementary Material
Acknowledgments
The authors thank all donors for their donations, and they are grateful to Hong-Sheng Wang (Sun Yat-sen University) for sharing plasmids in the dm6ACRISPR system.
Supported by National Natural Science Foundation of China (82070974 to XC, 82271100 to QHL); Natural Science Foundation of Jiangsu Province (BK20231371 to XC); Jiangsu “333” Advanced Talent-training Project to XC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability Statement: The raw RNA-seq data reported in this study have been deposited in the GEO database under the accession number GSE247663.
Disclosure: Y. Wang, None; Y.-R. Zhang, None; Z.-Q. Ding, None; Y.-C. Zhang, None; R.-X. Sun, None; H.-J. Zhu, None; J.-N. Wang, None; B. Xu, None; P. Zhang, None; J.-D. Ji, None; Q.-H. Liu, None; X. Chen, None
References
- 1. Bressler NM. Age-related macular degeneration is the leading cause of blindness. JAMA. 2004; 291(15): 1900–1901. [DOI] [PubMed] [Google Scholar]
- 2. Jager RD, Mieler WF, Miller JW.. Age-related macular degeneration. N Engl J Med. 2008; 358(24): 2606–2617. [DOI] [PubMed] [Google Scholar]
- 3. Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY.. Age-related macular degeneration. Lancet. 2012; 379(9827): 1728–1738. [DOI] [PubMed] [Google Scholar]
- 4. Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005; 85(3): 845–881. [DOI] [PubMed] [Google Scholar]
- 5. Datta S, Cano M, Ebrahimi K, Wang L, Handa JT. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog Retin Eye Res. 2017; 60: 201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaarniranta K, Uusitalo H, Blasiak J, et al.. Mechanisms of mitochondrial dysfunction and their impact on age-related macular degeneration. Prog Retin Eye Res. 2020; 79: 100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jun S, Datta S, Wang L, Pegany R, Cano M, Handa JT.. The impact of lipids, lipid oxidation, and inflammation on AMD, and the potential role of miRNAs on lipid metabolism in the RPE. Exp Eye Res. 2019; 181: 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kopp F, Mendell JT.. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018; 172(3): 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karali M, Banfi S.. Non-coding RNAs in retinal development and function. Hum Genet. 2019; 138(8-9): 957–971. [DOI] [PubMed] [Google Scholar]
- 10. Blasiak J, Hyttinen JMT, Szczepanska J, Pawlowska E, Kaarniranta K. Potential of long non-coding RNAs in age-related macular degeneration. Int J Mol Sci. 2021; 22(17): 9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim H, Kim J, Ryu J. Noncoding RNAs as a novel approach to target retinopathy of prematurity. Front Pharmacol. 2022; 13: 1033341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hyttinen JMT, Blasiak J, Kaarniranta K.. Non-coding RNAs regulating mitochondrial functions and the oxidative stress response as putative targets against age-related macular degeneration (AMD). Int J Mol Sci. 2023; 24(3): 2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mercer TR, Munro T, Mattick JS.. The potential of long noncoding RNA therapies. Trends Pharmacol Sci. 2022; 43(4): 269–280. [DOI] [PubMed] [Google Scholar]
- 14. Stelzer Y, Sagi I, Yanuka O, Eiges R, Benvenisty N.. The noncoding RNA IPW regulates the imprinted DLK1-DIO3 locus in an induced pluripotent stem cell model of Prader-Willi syndrome. Nat Genet. 2014; 46(6): 551–557. [DOI] [PubMed] [Google Scholar]
- 15. Deshpande RP, Sharma S, Liu Y, et al.. LncRNA IPW inhibits growth of ductal carcinoma in situ by downregulating ID2 through miR-29c. Breast Cancer Res. 2022; 24(1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Z, Jiang H, Zhao R, Quan Q, Huang X.. Long noncoding RNA IPW Is a novel diagnostic and predictive biomarker in lung adenocarcinoma. Genet Test Mol Biomarkers. 2023; 27(1): 18–26. [DOI] [PubMed] [Google Scholar]
- 17. Yang TJ, Yao MD, Sun YN, Li XM, Jiang Q, Yan B.. Suppression of choroidal neovascularization by silencing of long non-coding RNA IPW. Aging (Albany NY). 2021; 13(7): 10584–10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gamm DM, Melvan JN, Shearer RL, et al.. A novel serum-free method for culturing human prenatal retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2008; 49(2): 788–799. [DOI] [PubMed] [Google Scholar]
- 19. Osakada F, Jin ZB, Hirami Y, et al.. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci. 2009; 122(Pt 17): 3169–3179. [DOI] [PubMed] [Google Scholar]
- 20. Chen X, Jiang C, Sun R, Yang D, Liu Q.. Circular noncoding RNA NR3C1 acts as a miR-382-5p sponge to protect RPE functions via regulating PTEN/AKT/mTOR signaling pathway. Mol Ther. 2020; 28(3): 929–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chan CM, Huang DY, Sekar P, Hsu SH, Lin WW.. Reactive oxygen species-dependent mitochondrial dynamics and autophagy confer protective effects in retinal pigment epithelial cells against sodium iodate-induced cell death. J Biomed Sci. 2019; 26(1): 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shanmuganathan S, Angayarkanni N.. Chebulagic acid Chebulinic acid and Gallic acid, the active principles of Triphala, inhibit TNFalpha induced pro-angiogenic and pro-inflammatory activities in retinal capillary endothelial cells by inhibiting p38, ERK and NFkB phosphorylation. Vascul Pharmacol. 2018; 108: 23–35. [DOI] [PubMed] [Google Scholar]
- 23. Zhao C, Yasumura D, Li X, et al.. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest. 2011; 121(1): 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X, Wu Z, Fu X, Han W.. lncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014; 762: 1–21. [DOI] [PubMed] [Google Scholar]
- 25. Zou H, Shan C, Ma L, Liu J, Yang N, Zhao J. Polarity and epithelial-mesenchymal transition of retinal pigment epithelial cells in proliferative vitreoretinopathy. PeerJ. 2020; 8: e10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamiya S, Kaplan HJ.. Role of epithelial-mesenchymal transition in proliferative vitreoretinopathy. Exp Eye Res. 2016; 142: 26–31. [DOI] [PubMed] [Google Scholar]
- 27. Yao J, Bi HE, Sheng Y, et al.. Ultraviolet (UV) and hydrogen peroxide activate ceramide-ER stress-AMPK signaling axis to promote retinal pigment epithelium (RPE) cell apoptosis. Int J Mol Sci. 2013; 14(5): 10355–10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cusenza VY, Tameni A, Neri A, Frazzi R.. The lncRNA epigenetics: the significance of m6A and m5C lncRNA modifications in cancer. Front Oncol. 2023; 13: 1063636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J, Chen Z, Chen F, et al.. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res. 2020; 48(10): 5684–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feng C, Liu Y, Wang G, et al.. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014; 289(17): 11571–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen L, Zhang C, Ma W, Huang J, Zhao Y, Liu H.. METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability. Nucleic Acids Res. 2022; 50(20): 11619–11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen ZH, Chen TQ, Zeng ZC, et al.. Nuclear export of chimeric mRNAs depends on an lncRNA-triggered autoregulatory loop in blood malignancies. Cell Death Dis. 2020; 11(7): 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen X, Wang Y, Wang JN, et al.. m(6)A modification of circSPECC1 suppresses RPE oxidative damage and maintains retinal homeostasis. Cell Rep. 2022; 41(7): 111671. [DOI] [PubMed] [Google Scholar]
- 34. Sun HJ, Zhang FF, Xiao Q, Xu J, Zhu LJ.. lncRNA MEG3, acting as a ceRNA, modulates RPE differentiation through the miR-7-5p/Pax6 axis. Biochem Genet. 2021; 59(6): 1617–1630. [DOI] [PubMed] [Google Scholar]
- 35. Luo R, Jin H, Li L, Hu YX, Xiao F.. Long noncoding RNA MEG3 inhibits apoptosis of retinal pigment epithelium cells induced by high glucose via the miR-93/Nrf2 axis. Am J Pathol. 2020; 190(9): 1813–1822. [DOI] [PubMed] [Google Scholar]
- 36. Jing X, Han J, Zhang J, et al.. Long non-coding RNA MEG3 promotes cisplatin-induced nephrotoxicity through regulating AKT/TSC/mTOR-mediated autophagy. Int J Biol Sci. 2021; 17(14): 3968–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ye M, Lu H, Tang W, et al.. Downregulation of MEG3 promotes neuroblastoma development through FOXO1-mediated autophagy and mTOR-mediated epithelial-mesenchymal transition. Int J Biol Sci. 2020; 16(15): 3050–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He Y, Dan Y, Gao X, Huang L, Lv H, Chen J.. DNMT1-mediated lncRNA MEG3 methylation accelerates endothelial-mesenchymal transition in diabetic retinopathy through the PI3K/Akt/mTOR signaling pathway. Am J Physiol Endocrinol Metab. 2021; 320(3): E598–E608. [DOI] [PubMed] [Google Scholar]
- 39. Xia W, Li C, Chen Q, et al.. Intravenous route to choroidal neovascularization by macrophage-disguised nanocarriers for mTOR modulation. Acta Pharm Sin B. 2022; 12(5): 2506–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y, Fung NSK, Lam WC, Lo ACY.. mTOR signalling pathway: a potential therapeutic target for ocular neurodegenerative diseases. Antioxidants (Basel). 2022; 11(7): 1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen X, Jiang C, Qin B, et al.. LncRNA ZNF503-AS1 promotes RPE differentiation by downregulating ZNF503 expression. Cell Death Dis. 2017; 8(9): e3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen X, Sun R, Yang D, Jiang C, Liu Q.. LINC00167 regulates RPE differentiation by targeting the miR-203a-3p/SOCS3 Axis. Mol Ther Nucleic Acids. 2020; 19: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hirsch L, Nazari H, Sreekumar PG, et al.. TGF-beta2 secretion from RPE decreases with polarization and becomes apically oriented. Cytokine. 2015; 71(2): 394–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rizzolo LJ. Polarity and the development of the outer blood-retinal barrier. Histol Histopathol. 1997; 12(4): 1057–1067. [PubMed] [Google Scholar]
- 45. Tang SJ, You GR, Chang JT, Cheng AJ.. Systematic analysis and identification of dysregulated panel lncRNAs contributing to poor prognosis in head-neck cancer. Front Oncol. 2021; 11: 731752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luzon-Toro B, Villalba-Benito L, Fernandez RM, Torroglosa A, Antinolo G, Borrego S.. RMRP, RMST, FTX and IPW: novel potential long non-coding RNAs in medullary thyroid cancer. Orphanet J Rare Dis. 2021; 16(1): 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen Y, Lin Y, Shu Y, He J, Gao W.. Interaction between N6-methyladenosine (m6A) modification and noncoding RNAs in cancer. Mol Cancer. 2020; 19(1): 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kumari R, Ranjan P, Suleiman ZG, et al.. mRNA modifications in cardiovascular biology and disease: with a focus on m6A modification. Cardiovasc Res. 2022; 118(7): 1680–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y, Chen Y, Liang J, et al.. METTL3-mediated m6A modification of HMGA2 mRNA promotes subretinal fibrosis and epithelial-mesenchymal transition. J Mol Cell Biol. 2023; 15: mjad005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao B, Huang J, Lou X, et al.. Endothelial CYP2J2 overexpression restores the BRB via METTL3-mediated ANXA1 upregulation. FASEB J. 2022; 36(11): e22619. [DOI] [PubMed] [Google Scholar]
- 51. Hu Y, Chen J, Wang Y, et al.. Fat mass and obesity-associated protein alleviates Abeta(1-40) induced retinal pigment epithelial cells degeneration via PKA/CREB signaling pathway. Cell Biol Int. 2023; 47(3): 584–597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.