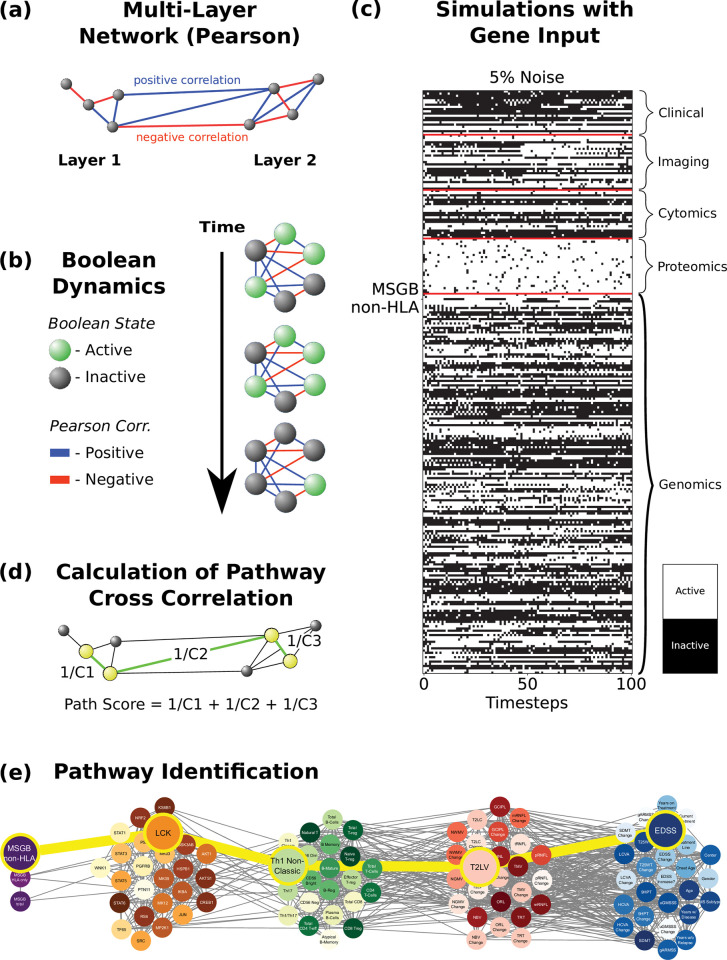
Fig 3. Dynamic network analysis: identification of gene-protein-cell paths.
(a) Networks are constructed using all five layers. The nodes are the same as in the networks above (Fig 1), but now the edges are defined by the Pearson correlation, where the weights represent the Pearson coefficient, which can be either positive or negative. (b) Boolean dynamics are applied to the networks, where the activation state of the nodes changes based on the total sum of the edge weights of its direct neighbors (considering the signs of the connections). (c) Boolean simulations are run where the various nodes, in the example MSGB non-HLA, are used as the input signal, and the simulation was run with 5% noise (see Methods for noise analysis). (d) The cross-correlation coefficient (Cn) is calculated between the signals for each pair of connected nodes. A path score is calculated for all possible paths, defined as the sum of the inverses of the cross-correlation coefficients between all pairs of consecutive nodes constituting a given path. (e) Finally, a path is identified by using a shortest path algorithm which is based on its path score (see Methods).