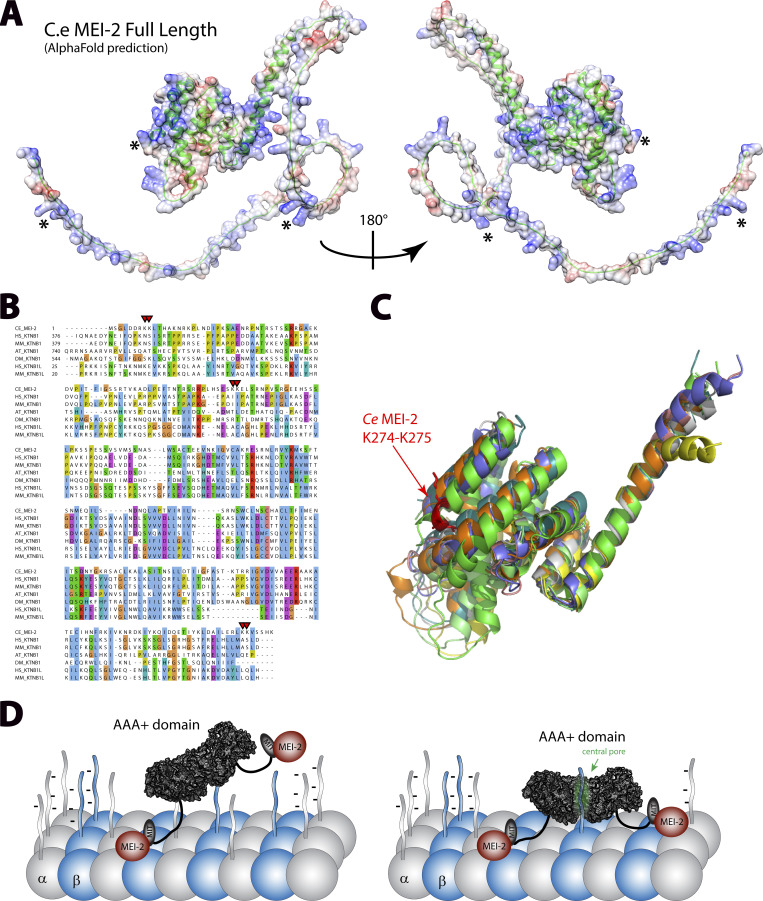
Figure 6.
p80 structural alignment and Katanin•MT interacting model. (A) Structural model of MEI-2 obtained using AlphaFold2 prediction website. The carbon backbone is represented using cartoon mode (in green), and the surface color shows the electrostatic potential calculated using chimera electrostatic potential Coulombic function. * corresponds to the locations of KK pair residues identified in this study. (B) Protein sequences alignment of p80 and p80-like Katanin subunits from different species. CE: C. elegans; HS: H. sapiens; MM: M. musculus; AT: A. thaliana; DM: D. melanogaster. (C) Structural alignment of the C-terminal part of p80 and p80-like Katanin subunits from different species. Except for MM KATNB1, which is the X-ray structure obtained in complex with p60 fragment (PDB: 5LB7) (Jiang et al., 2017), structures shown are alpha-fold prediction aligned on MEI-2 structural prediction. The region shown is limited to the “structured region” of MEI-2 (aa 105–280). Single-protein structures are presented in Fig. S4. (D) Working model of microtubule binding of Katanin essential for microtubule severing activity.