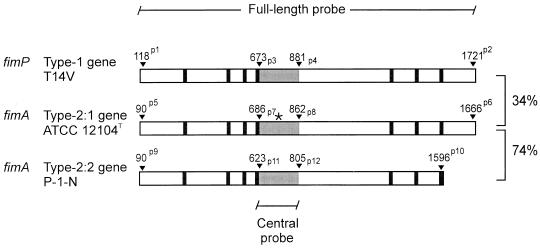
FIG. 1.
Schematic illustration of the fimP and fimA fimbrial subunit genes from A. naeslundii. DNA probes representing full-length and central (gray area) gene segments specific for different adhesin specificities were PCR amplified by using synthetic oligonucleotide primers specific for fimP from A. naeslundii T14V (genospecies 2), fimA from ATCC 12104T (genospecies 1), and fimA from P-1-N (genospecies 2, strain CCUG 33910). The nucleotide positions of all primers are indicated by arrowheads and labeled p1 through p12 (see Materials and Methods). Primers set as superscripts to position numbers were used to amplify full-length gene segments; primers set as subscripts to position numbers were used to amplify central gene segments. The central DNA probes were generated from a region of the gene sequences having comparably low homology and being devoid of highly conserved proline-containing segments. The overall nucleotide sequence identities between the three fimbrial subunit genes are indicated on the right. An asterisk marks the position of the BamHI cleavage site in the type 2:1 fimbrial subunit gene.