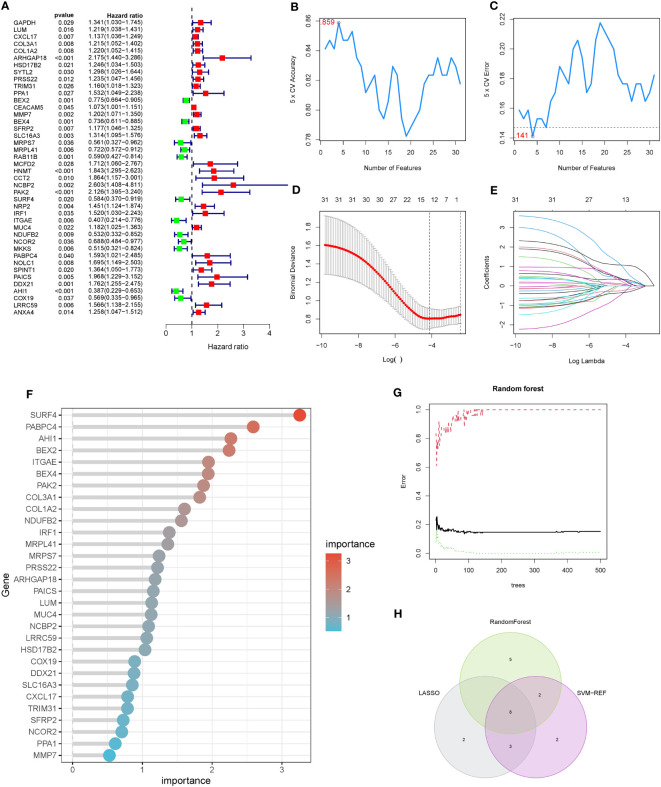
Figure 3.
COX regression model combined with machine learning algorithms for the selection of pancreatic cancer liver metastasis-related genes. (A) Results of single-factor COX regression analysis on 139 genes in the TCGA cohort. Hazard ratio values less than 1 indicate protective factors for the prognosis of pancreatic cancer patients, while values greater than 1 indicate risk factors. (B, C) SVM-RFE machine learning feature selection. The top 15 genes were extracted based on the average ranking from ten-fold cross-validation, including COL1A2, PRSS22, PAK2, SURF4, IRF1, PABPC4, AHI1, and ANXA4. (D, E) LASSO regression model feature selection. Thirteen genes were extracted based on the selection of lambda.min, including COL1A2, PRSS22, PAK2, SURF4, IRF1, PABPC4, AHI1, and ANXA4. (F, G) Feature selection using random forest analysis. The top 15 genes were extracted based on the ranking of gene importance, including COL1A2, PRSS22, PAK2, SURF4, IRF1, PABPC4, AHI1, and ANXA4. (H) Intersection of the gene sets obtained from the three machine learning methods, resulting in a final set of 8 genes.