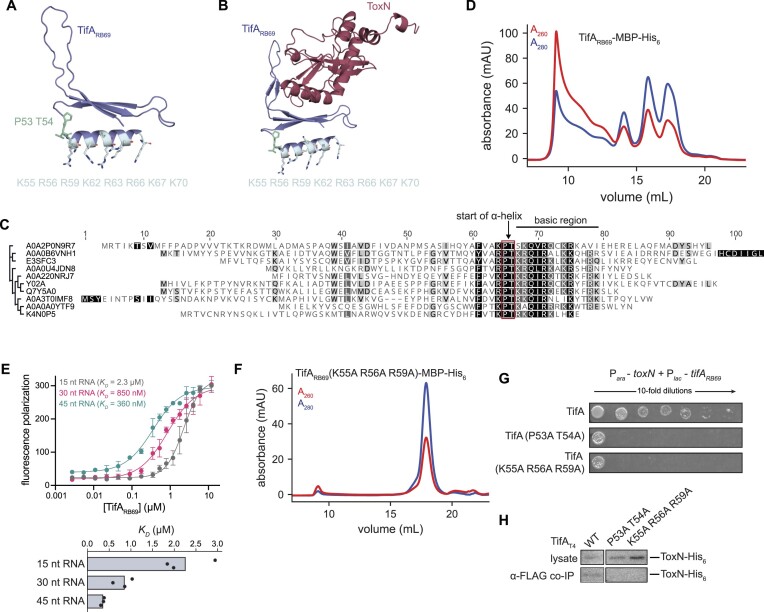
Figure 5.
TifA is an RNA-binding protein. (A) AlphaFold model for TifARB69 with residues capping the predicted C-terminal α-helix (P53 T54) and basic residues on the surface of the predicted α-helix indicated. (B) AlphaFold model for the ToxN-TifARB69 complex with residues capping the predicted C-terminal α-helix (P53 T54) and basic residues on the surface of the predicted α-helix indicated. (C) Sequence alignment for 10 TifA homologs from different phylogenetic clusters indicating the highly-conserved PT motif, immediately followed by a cluster of basic residues. UniProtKB accession numbers for the TifA homologs in the alignment are, from top to bottom, as follows: A0A2P0N9R7; A0A0B6VNH1; E3SFC3; A0A0U4JDN8; A0A220NRJ7; Y02A; Q7Y5A0; A0A3T0IMF8; A0A0A0YTF9; K4N0P5. (D) Size-exclusion chromatogram of purified TifARB69-MBP-His6 run on a Superose 6 Increase 10/300 GL column. (E) Fluorescence polarization assay of increasing concentrations of synthesized TifARB69 incubated with several fluorescently-labeled RNA species (lengths indicated). RNA sequences were derived from the region of the E. coli transcript artJ that is cleaved by ToxN; see Supplementary Table S5 for sequences. (F) Size-exclusion chromatogram of purified TifARB69 (K55A R56A R59A)-MBP-His6 run on a Superose 6 Increase 10/300 GL column. (G) Serial dilutions of E. coli cells ectopically expressing toxN and wild-type or RNA-binding deficient tifARB69. (H) Western blot of ToxN-His6 in cell lysate and following co-immunoprecipitation with TifAT4-FLAG (wild-type or RNA-binding deficient) in T4-infected cells producing TifAT4 and ToxN. Note that the TifAT4 wild-type control is same as in Figure 3E because both experiments were run on the same blot. Also see Supplementary Figure S6.