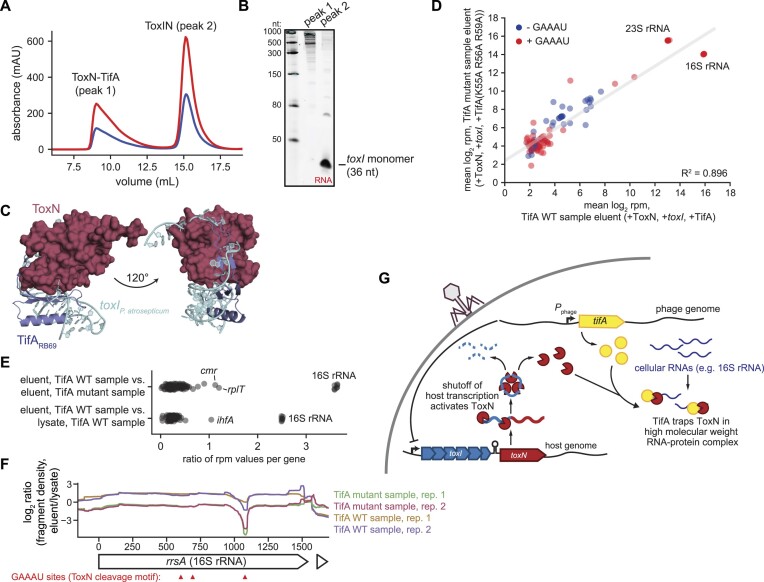
Figure 6.
ToxN co-purifies with the 16S rRNA in the presence of TifA. (A) Size-exclusion chromatogram of ToxN co-purified with toxI and TifA run a Superose 6 Increase 10/300 GL column. (B) 6% urea–PAGE gel analyzing the RNA content of the two major peaks in (A). (C) AlphaFold model for the ToxN-TifARB69 complex, with P. atrosepticum toxI (PDB: 2xdd) modelled into the complex. (D) Scatter plot comparing the mean summed RNA-seq counts (log2 RPM) for individual genes in the TifA WT sample eluent (+ToxN, +toxI, +TifARB69) and the TifA mutant sample eluent (+ToxN, +toxI, +TifARB69 (K55A R56A R59A)) (two biological replicates each). Genes with RPM values greater than or equal to 3 in all sequenced libraries (lysates and eluents) are shown. The gray line indicates the linear regression fit for the data. (E) Scatter plots showing the ratio of mean RPM values for individual transcripts in the samples indicated. (F) Plot of log2 (ratio, fragment density in eluent versus lysate) across the 16S rRNA locus rrsA for both biological replicates of the TifA WT and TifA mutant samples. Sites containing the ToxN cleavage motif (GAAAU) are indicated with red arrows. (G) Proposed model for TifA inhibition of ToxN. Following phage infection, shutoff of host transcription, including of the toxIN locus, leads to degradation of toxI and release of ToxN. TifA then interacts with activated ToxN to prevent degradation of cellular transcripts, likely by recruiting and/or sequestering ToxN to a subset of cellular RNAs, in particular the 16S rRNA, such that ToxN can no longer cleave phage transcripts. Also see Supplementary Figure S7.