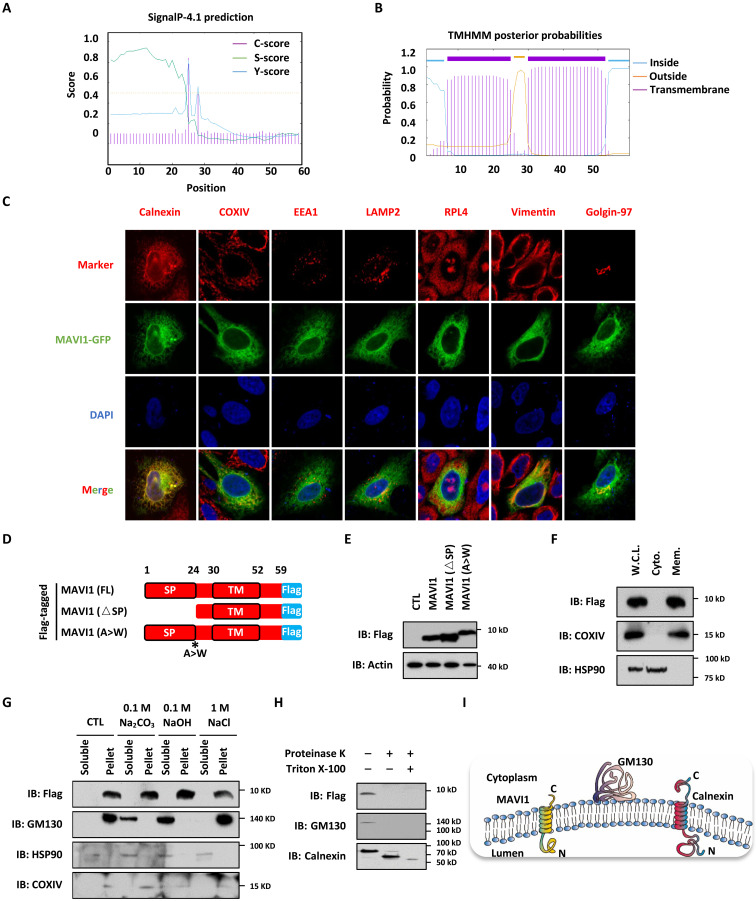
Fig. 4. MAVI1 is an ER membrane protein.
(A and B) The SP (1 to 24 amino acids) (A) and TM domain (30 to 52 aa) (B) in MAVI1 predicted by SignaIP-4.1and TMHMM website, respectively, are shown. (C) HEK293 cells transfected with GFP-tagged MAVI1 were stained with Calnexin (ER maker), COXIV (mitochondria maker), EEA1 (endosome maker), LAMP2 (lysosome maker), RPL4 (ribosome maker), Vimentin (cytoskeleton maker), or Golgin-97 (Golgi apparatus marker) (Red). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (D) Schematic representation of Flag-tagged, full length (FL) MAVI1, SP-deleted (△SP) MAVI1, and MAVI1 mutant with the SP cleavage site mutated (A > W) is shown. (E) HEK293 cells transfected with empty vector (CTL) or expression vectors as described in (D) were subjected to IB analysis. (F) HEK293 cells stably expressing Flag-tagged MAVI1 were subjected to cellular fraction, followed by IB analysis. W.C.L., whole cell lysate; Cyto., cytoplasmic fraction; Mem., membrane fraction. (G) HEK293 cells stably expressing Flag-tagged MAVI1 were subjected to membrane interaction assay, and the supernatant was treated with or without 0.1 M Na2CO3, 0.1 M NaOH, or 1 M NaCl. The resultant soluble solution and pellet were subjected to IB analysis. (H) HEK293 cells stably expressing Flag-tagged MAVI1 were subjected to cellular fractionation, and membrane fractions were treated with or without proteinase K in the presence or absence of Triton X-100, followed by IB analysis. (I) Membrane topology of calnexin, GM130, and MAVI1 proteins is shown.