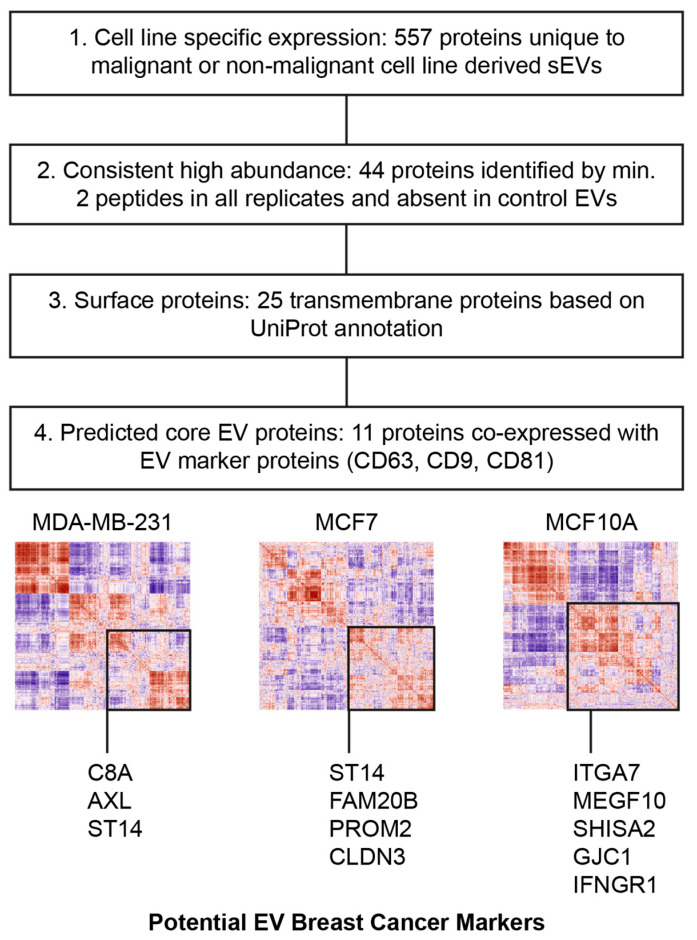
Figure 3.
Biomarker selection strategy. 1. Proteins uniquely identified in sEVs for each cell and common to MDA-MB-231 and MCF7 were selected. 2. Abundance by at least 2 peptides in all replicates and absence of any peptides in control cell lines were required. 3. Proteins were classified by UniProt subcellular locations to identify membrane proteins. 4. Only proteins with predicted co-localization with classical EV markers based on correlation matrices were used as potential biomarkers.