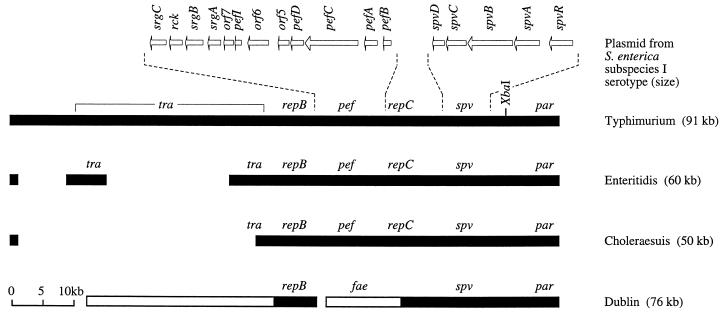
FIG. 3.
Linear genetic maps of virulence plasmids of S. enterica subspecies I serotypes. Plasmids were linearized at their conserved par (partitioning) region (29, 113, 114). Regions of homology shared among plasmids are shown as closed bars, which are positioned according to their locations on the S. enterica serotype Typhimurium virulence plasmid. The distribution among virulence plasmids of spv (Salmonella plasmid virulence) (6, 66, 122), repC (replicon C) (113), pef (plasmid encoded fimbriae) (12, 47), repB (replicon B) (113), and the tra (conjugative plasmid transfer) region (66, 101) have been reported recently. Areas of the plasmid of S. enterica serotype Dublin that are not present in S. enterica serotype Typhimurium have been described previously and are shown as open bars (68, 80). The presence on the S. enterica serotype Dublin plasmid of sequences with homology to feaH and feaI, two genes involved in biosynthesis of K88 fimbriae in E. coli, have been reported by Barrow and coworkers (105). The location and size of deletions that may account for the smaller size of plasmids from S. enterica serotypes Choleraesuis and Enteritidis relative to the S. enterica serotype Typhimurium plasmid are reported elsewhere (25, 80, 101). Dashed lines indicate the position of two DNA regions, namely the pef and spv operons, which are shown in more detail above the S. enterica serotype Typhimurium plasmid (1, 47, 52). The positions of genes are indicated by arrows.