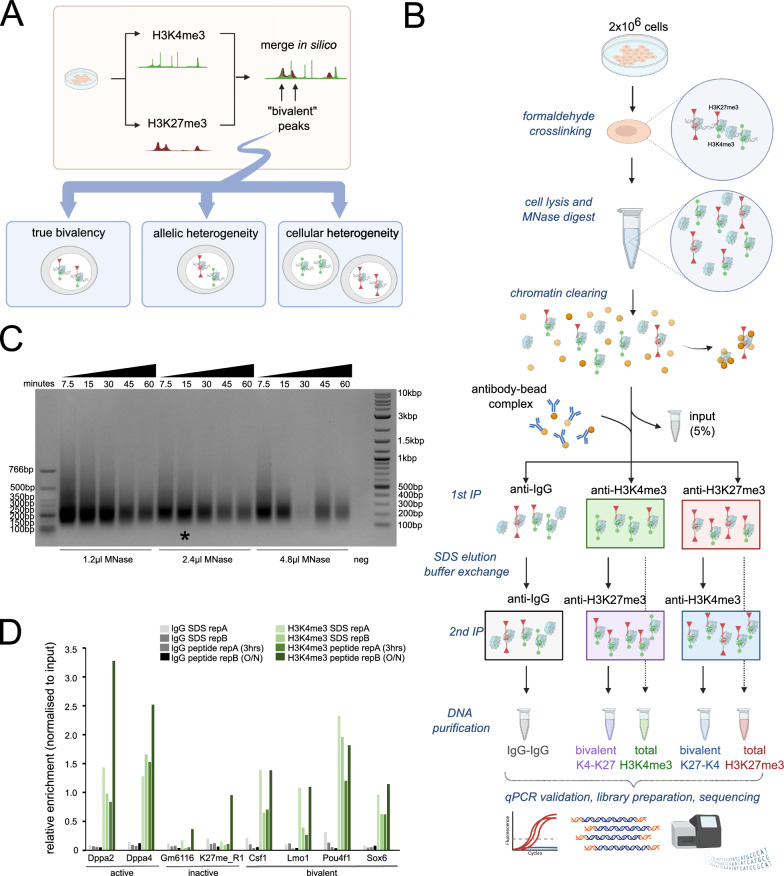
Fig. 1.
Development of an optimised ChIP-reChIP protocol to robustly measure bivalent chromatin. A Potential limitations in using independent total H3K4me3 (green, circles) and total H3K27me3 (red, triangles) datasets in distinguishing bone-fide bivalent chromatin, where the two marks occur on the same nucleosome, from allelic and cellular heterogeneity. B overview of sequential ChIP-reChIP protocol, see methods for detailed description. C Agarose gel showing chromatin digested with 1.2μl, 2.4μl or 4.8μl MNase for different amounts of time (7.5 min to 60 min). 2 million cells were used per condition. Negative control did not have any MNase added. Star denotes final condition used in protocol. D Single H3K4me3 (green) and IgG control (grey) ChIP-qPCR analysis comparing SDS-based elution (light) from peptide elution (dark). Two active H3K4me3-only (Dppa2, Dppa4), two inactive H3K27me3-only (Gm6116, K27me_R1) and four bivalently marked loci (Csf1, Lmo1, Pou4f1, Sox6) were analyzed. Enrichment values normalised to input are shown