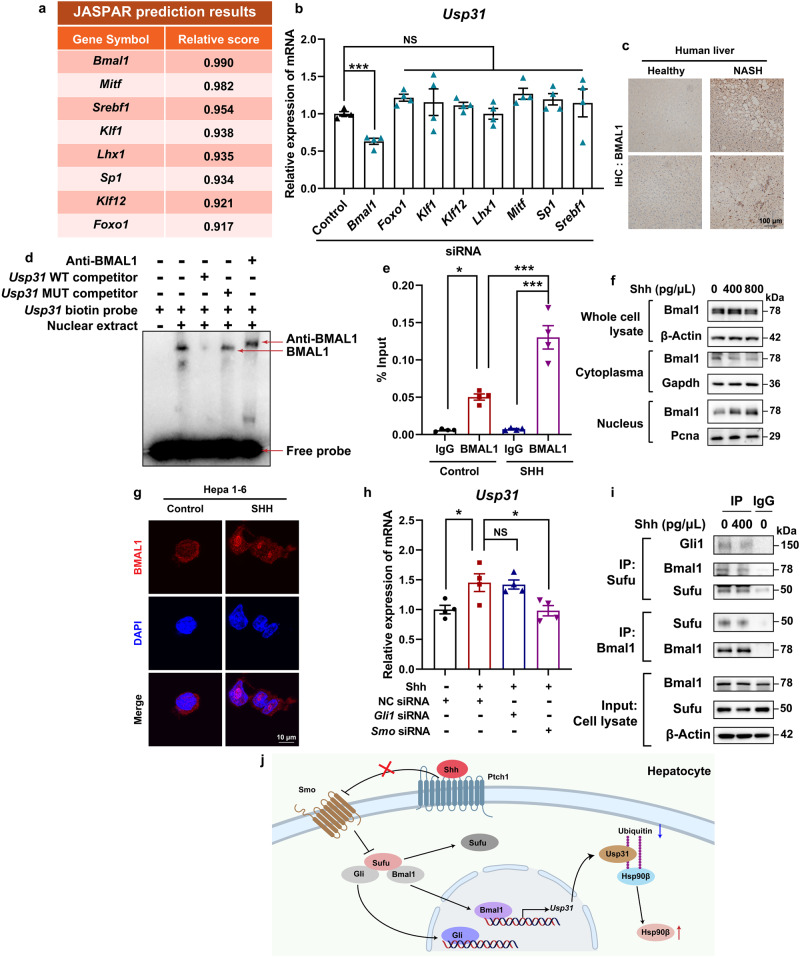
Fig. 5. BMAL1 is the transcription factors of Usp31.
a The transcription factors (TFs) of Usp31 were predicted using JASPAR. b Each of the predicted Usp31 TFs was knocked down by corresponding siRNA, the expression of Usp31 was then analyzed by qRT-PCR (Control siRNA versus Bmal1 siRNA, P = 0.0003, versus Foxo1 siRNA, P = 0.53, versus Klf1 siRNA, P = 0.82, versus Klf12 siRNA, P = 0.956, versus Lhx1 siRNA, P = 0.99, versus Mitf siRNA, P = 0.31, versus Sp1 siRNA, P = 0.64, versus Srebf1 siRNA, P = 0.86). c Representative immunohistochemistry staining of BMAL1 in the livers of healthy donors and NAFLD patients. d The EMSA assays were carried out with nuclear fraction of Hepa 1–6 cells. e Hepa 1–6 cells were treated with 400 pg/μL Shh for 24 h, the binding of BMAL1 to the Usp31 promoter region was analyzed by ChIP analysis (Control+IgG versus Control+BMAL1, P = 0.001, Control+BMAL1 versus SHH + BMAL1, P = 4.3E-05, SHH+IgG versus SHH + BMAL1, P = 5.1E-07). f Hepa 1–6 cells were treated with 400 pg/μL Shh for 24 h, cytoplasmic and nuclear BMAL1 were analyzed by western blot. g Hepa 1–6 cells were treated with 400 pg/μL Shh for 24 h, the cellular Bmal1 distribution was analyzed by immunofluorescence staining. h Hepa 1–6 cells were treated indicated siRNAs for 48 h, and then incubated with 400 pg/μL Shh for 24 h, the gene expression of Usp31 was analyzed by qRT-PCR (Shh+NC siRNA versus NC siRNA, P = 0.020, versus Shh+Gli1 siRNA, P = 0.99, versus Shh+Smo siRNA, P = 0.016). i Hepa 1–6 cells were treated with 400 pg/μL Shh for 24 h, the interaction between Bmal1/Gli1 and Sufu was analyzed by immunoprecipitation and western blot. j The signaling pathway through which Shh promotes Hsp90β deubiquitylation by Usp31. Data are presented as mean ± SEM. n = 6 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001, NS no significant difference, one-way ANOVA. Source data are provided in the Source Data file.