Fig. 1 |. Co-diversification of gut microbiota with primate species.
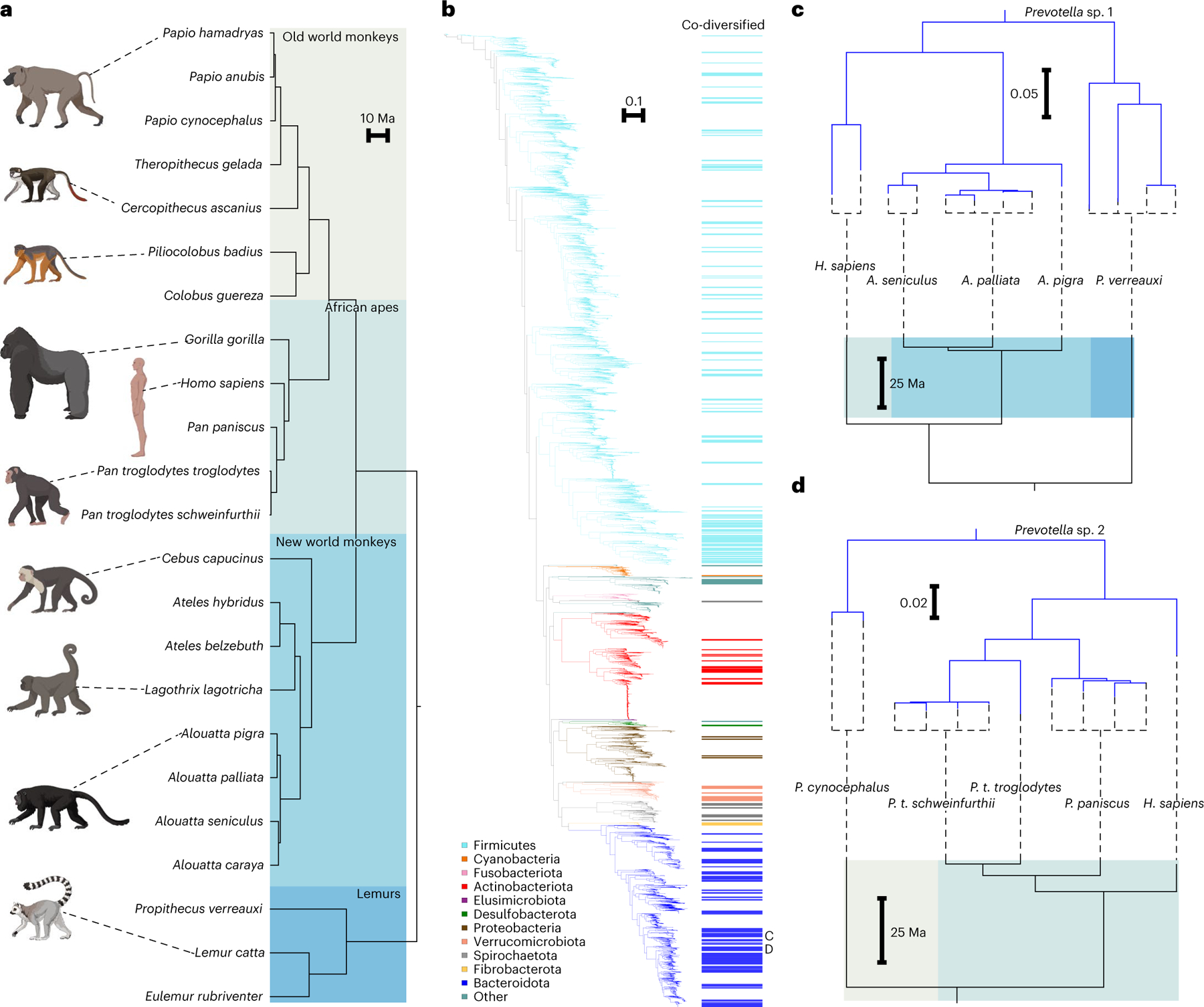
a, Phylogeny shows relationships among primate species from which gut bacterial genomes were derived. Shading delineates Old World monkeys, New World monkeys, African apes and lemurs. b, Phylogeny shows relationships among 9,460 high-quality gut bacterial genomes assembled from the primate gut microbiota. Coloured bars to the right of the phylogeny mark clades showing significant evidence of co-diversification with primate species (Mantel r > 0.75; non-parametric permutation test P < 0.01). Colours of branches and bars correspond to bacterial phyla. Scale indicates amino-acid substitutions per site. Letters C and D indicate clades highlighted in c and d, respectively. c,d, Tanglegrams show examples of correspondence between topologies of bacterial (top of each panel) and host-species (bottom of each panel) phylogenies for two co-diversifying clades of unclassified Prevotella species.
