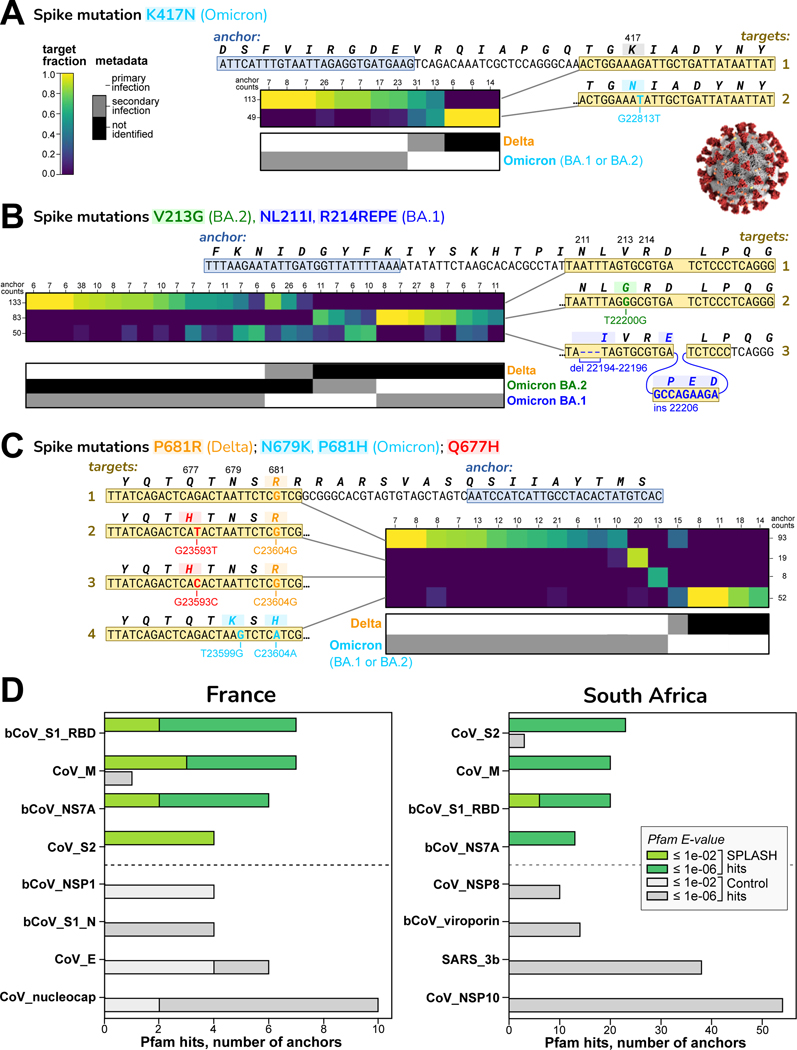
Figure 2. SPLASH identifies strain-defining and other variation in SARS-Cov2.
In A-C, sets of targets that distinguish SARS-CoV-2 strains are shown; all are in the spike protein (S) gene. Each heatmap has columns for different samples (patients) and rows for different targets; the coloring indicates the fraction of that target observed in that patient. Summary anchor counts are given for rows and columns. Also shown is a map of the categorical metadata of what strains (primary and secondary) were identified in each patient in the original study; this data was not used by SPLASH, but there is evident agreement between the heatmap and the metadata strain assignment. Binomial -value bounds were computed per STAR Methods.
A. Distinguishing at the major strain level: target 1 (no mutation) matches Delta; target 2 contains K417N, found in all Omicron (both BA.1 and BA.2 sub-strains); two patients co-infected with Delta and Omicron show both targets. ().
B. Distinguishing at the sub-strain level: target 1 (no mutation) matches Delta; target 2 with V213G is specific for BA.2; target 3 with a deletion (NL211I) and insertion (R214REPE) is specific for BA.1. ()
C. Distinguishing non-strain related mutations: target 1 has P681R, Delta specific; targets 2 and 3 encode Q677H (by different mutations) and P681R; target 4 has N679K and P681H, Omicron specific. ()
D. Protein domain profiling in SARS-CoV-2. The top four and bottom four Pfam protein domain hits are shown. S1 Receptor binding domain (RBD) and S2 domain show high variation by SPLASH in both datasets. Other Pfam abbreviations: bCoV = beta-coronavirus; CoV = coronavirus, nucleocap = nucleocapsid N = N-terminal domain, SARS = Severe acute respiratory syndrome coronavirus, M, NS7A, NSP1, NSP8, 3b, NSP10 = viral proteins.