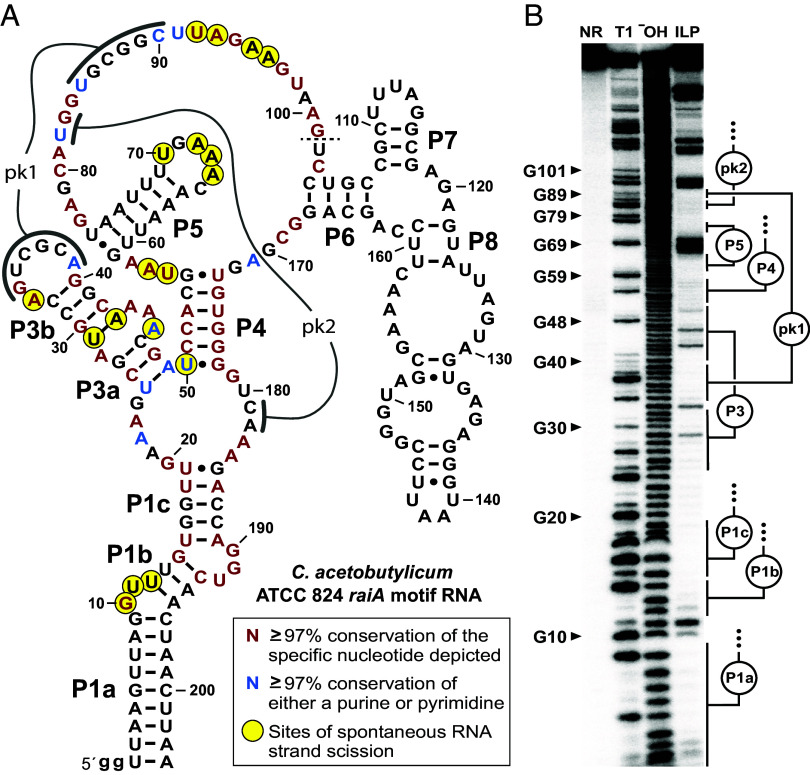
Fig. 3.
Evidence that raiA motif RNAs form complex-folded structures. (A) Sequence and secondary structure model for the raiA motif representative from C. acetobutylicum. Predicted base-paired stems are labeled P1 through P8. Nucleotides involved in predicted pseudoknots (pk1 and pk2) are also identified. Colored nucleotides match highly conserved nucleotides of the consensus model as indicated. Sites of spontaneous RNA strand scission were derived from in-line probing assay data depicted in (B). The dashed line at position 101 indicates that the in-line probing data was not mapped beyond this point. Lowercase “g” letters indicate nucleotides added to the construct to facilitate preparation by in vitro transcription. (B) Autoradiogram of a denaturing polyacrylamide gel electrophoresis (PAGE) separation of the products of an in-line probing reaction using trace amounts of 5′ 32P-labeled C. acetobutylicum raiA motif RNA. RNAs were not reacted (NR), subjected to partial digestion with RNase T1 (T1) or alkaline conditions (−OH), or were incubated under in-line probing conditions at room temperature (~20 °C) for 72 h. Selected bands corresponding to products resulting from RNase T1 digestion (cleavage after G nucleotides) are labeled accordingly.