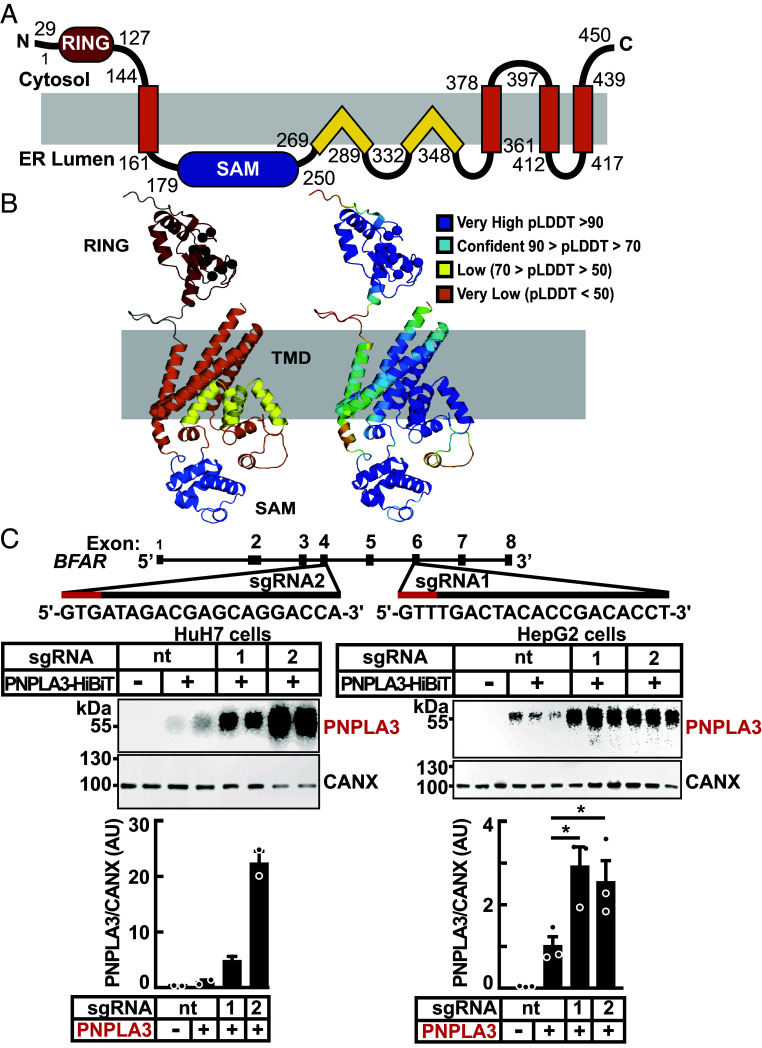
Fig. 3.
Domain organization, structural model, and inactivation of BFAR in cultured hepatocytes. (A) Schematic of the human BFAR domain structure model is colored: RING (red), sterile alpha motif (SAM) (blue), and transmembrane domains with TMH (orange) and re-entrant TM helices (yellow). (B) AlphaFold structure model of BFAR colored and labeled by domains as in (A), with zinc coordinating residues from the ring domain indicated (black sphere for the C-alpha position) and according to pLDDT confidence (Right, scale in legend) colored from very high (blue) to very low (orange). (C) Inactivation of BFAR using CRISPR/Cas9 in cultured hepatocytes results in increased levels of PNPLA3. Schematic of single guide (sg)RNAs used to inactivate BFAR is shown (Top). Two sgRNAs targeting BFAR exons 4 (sgRNA2) and 6 (sgRNA1) were used to introduce frameshift mutations that prematurely terminate the protein at residues 209 and 361, respectively. Red lines denote the Protospacer Adjacent Motif (PAM) sequence. HuH7 cells were transduced with LentiCRISPRv2 containing sgRNA1, sgRNA2, or with a sgRNA with no cognate sequence in the human genome (nontargeting, nt) and cell lines were established from single clones (Materials and Methods). The BFAR−/− HuH7 and HepG2 cells were transfected with PNPLA3-HiBiT (2.5 ng/well) and after 48 h, PNPLA3 levels were measured in cell extracts using the NanoBiT blotting assay. Luminescent signals were quantified using LI-COR, normalized to CANX levels and expressed as arbitrary units (AUs). Values represent means ± SEM. Levels were compared using a Student’s t test. *P < 0.05. The experiments were repeated twice and the results were similar.