Fig. 2.
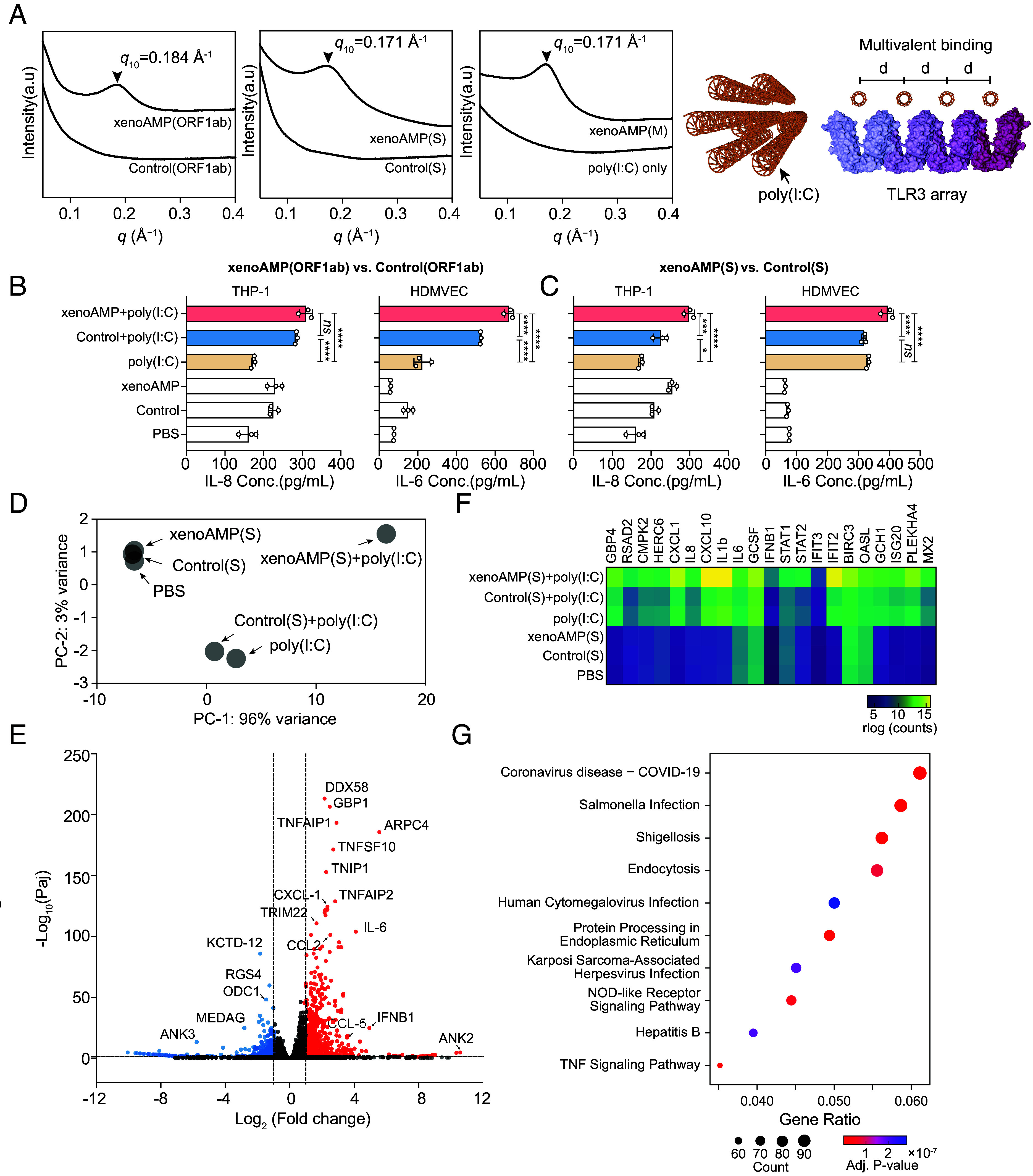
XenoAMPs from SARS-CoV-2 but not homologs from HCoV-OC43 organize poly(I:C) into ordered complexes that amplify immune responses. (A) SAXS data show that SARS-CoV-2 derived complexes exhibit a liquid crystalline structure that can amplify activation of TLR3, whereas HCoV-OC43 complexes do not. (B and C) Cytokine release from THP-1 monocytes and human dermal microvascular endothelial cells (HDMVEC) that are stimulated with SARS-CoV-2 xenoAMP-poly(I:C) complexes or the HCoV-OC43 Control-poly(I:C) complex (n = 3). Data is presented as mean ± SE. Statistical analysis is performed using one-way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (D) Principal component analysis of the transcriptome profile in HDMVEC treated by xenoAMP(S)-poly(I:C) complexes, Control(S)-poly(I:C), poly(I:C) only and associated controls. (E) Differential gene expression in HDMVEC treated by xenoAMP(S)-poly(I:C) compared to the poly(I:C) treatment group. Significance (−Log10(Paj), y axis) is plotted against magnitude of gene expression (Log2(Fold change), x axis). Significantly upregulated and downregulated genes (Paj < 0.05, Log2(Fold change) > 1; Log2(Fold change) <−1) are highlighted in red and blue respectively. (F)Transcriptional change of immune-related genes in HDMVEC in different conditions. (G) By comparing all significantly upregulated genes (P < 0.05) to curated expression patterns from the KEGG database, the best match to the gene expression pattern induced by complexes with the xenoAMP(S) fragment is COVID-19 (P < 1 × 10−7).
