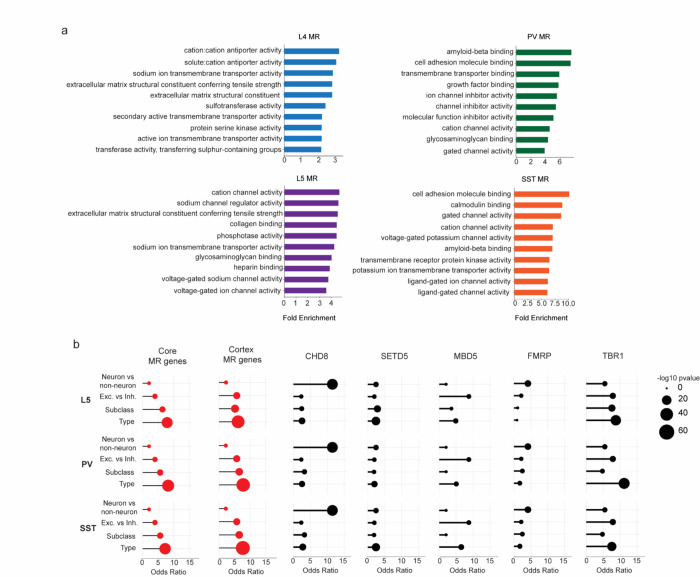
Extended Data Fig. 5. Functional annotation of subclass-defined MeCP2-repressed genes and their representation in other datasets.
a, Gene ontology of MeCP2-repressed (MR) genes in L4, L5, PV, and SST neurons. Top 10 terms for Molecular Function shown. b, Overlap of differentially expressed genes across L5, PV, and SST cellular hierarchy with core MeCP2-repressed genes, MeCP2-repressed genes previously identified in the cortex12, and dysregulated genes detected in other NDD mouse models. Note that each differential list labeled on the left for L5, PV and SST reflects the same neuron vs. non-neuron and excitatory vs. inhibitory lists, but the lists of differential genes at the subclass and type level reflect the genes that specifically define these differentiations in L5, PV or SST cells.
Analysis was performed on differentially expressed genes from INTACT RNA-seq analysis described in Figure 1. n=4 biological replicates per genotype per cell type, as well as the indicated published gene lists.