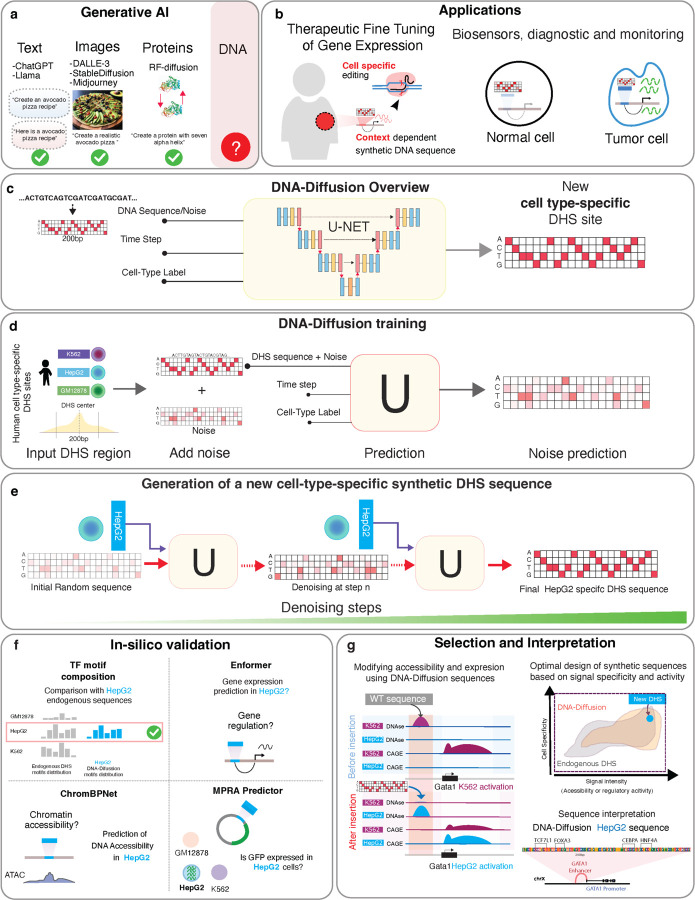
Figure 1.
a) Examples of generative AI models applied to language, image generation, and biological problems. b) Exploration of potential therapeutic applications based on synthetic cell-type specific sequences. c) Schematic overview of the DNA-Diffusion model. The model utilizes a U-Net architecture to generate new DNA sequences iteratively based on cell types presented in the training dataset. d) The DNA-Diffusion model was trained using unique DHS DNA sequences from different cell types, including K562, HepG2, and GM12878. The training involves transforming the endogenous DHS sequence into a hot-encoded format and introducing a fixed amount of standard normal noise. The trained U-Net uses the expected noise level (determined by the time step) and cell type information to predict and remove the added noise. This noise prediction process is repeated during training across the entire sequence dataset with varied noise intensities. Once trained, the U-Net can predict the initial noise added to the original DHS endogenous sequences, enabling the generation of new sequences specific to different cell types. e) To generate a new sequence given a cell-type label, a hot-encoded DNA matrix with random Gaussian noise is generated, and the U-Net iteratively denoises this matrix over 50 steps, progressively converging to a sequence that reflects the characteristics of the target cell type. f) Different in silico validations were utilized to evaluate the accessibility, regulatory activity, and motif composition of DNA-Diffusion and endogenous DHS regions. g) Framework developed for selecting and interpreting generated sequences based on cell-type signal specificity, intensity, or motif composition.