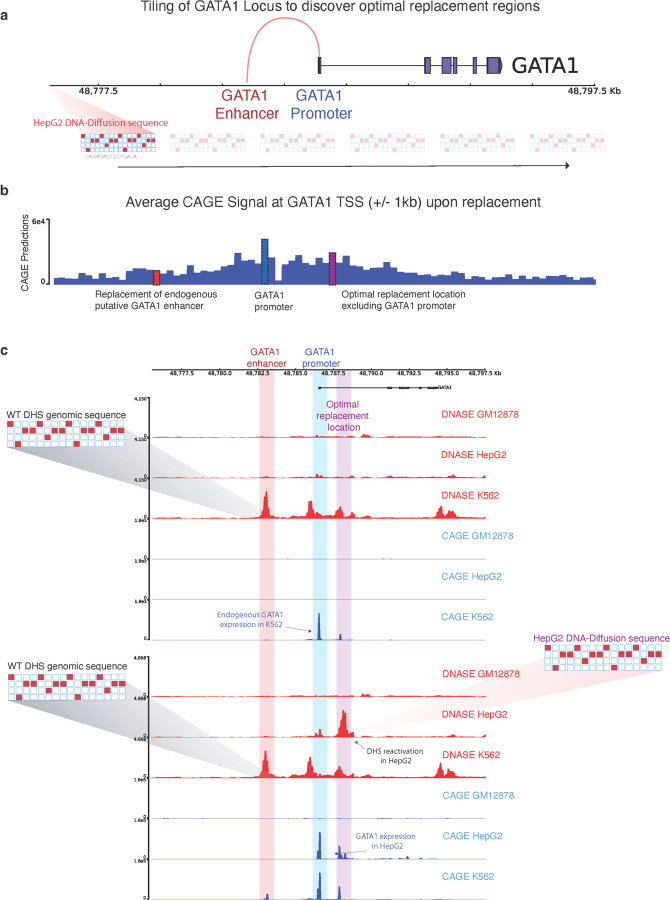
Figure 4.
a) Tiling approach for exploring the optimal location of replacement of a synthetic sequence to reactivate a gene of interest. b) The average CAGE predictions for the GATA1 TSS+/- 1KB are shown for each replacement of the same HepG2-specific sequence throughout the entire locus. Key regions with strong predicted functional effects on GATA1 reactivation are highlighted; the original GATA1 enhancer in K562 (red), the GATA1 promoter (blue), and the optimal replacement region discovered by the tiling procedure (purple). c) Top: DNase and CAGE activity predictions at the GATA1 locus based on the wild-type sequence. Bottom: post-insertion of a HepG2-optimized DNA-Diffusion sequence into the optimal replacement location, resulting in heightened chromatin accessibility and GATA1 expression in HepG2, maintenance of accessibility and GATA expression in K562 and no significant changes in GM12878.