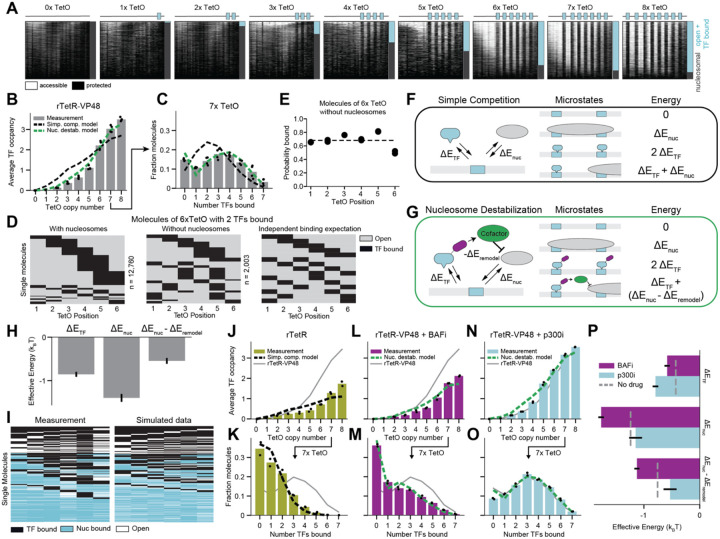
Figure 2: Thermodynamic models reveal rTetR-VP48 competes with nucleosomes with the aid of activation domains.
A) Summary plots of enhancer methylation across single molecules observed by SMF for enhancers with 0-8 TetO binding sites after 24 hours of 1,000 ng/ml dox induction. Each row represents methylation data for one molecule, as in 1F.
B) Observed average occupancy of rTetR-VP48 for enhancers with increasing numbers of TetO binding sites (four biological replicates) fit to a Simple Competition model (described in F, r2 = 0.84) or a Nucleosome Destabilization model (described in G, r2 = 0.99).
C) Observed occupancy distribution of rTetR-VP48 TFs for the enhancer with 7 TetO sites (four biological replicates) fit to a Simple Competition model (described in F, r2 = −0.72) or a Nucleosome Destabilization model (described in G, r2 = 0.79).
D) Observed distributions of occupied TetO sites on molecules of 6xTetO with 2 TFs bound with and without nucleosomes present and the predictions from a model that assumes independent binding.
E) Likelihood of distinct TetO binding sites of being occupied in nucleosome-free molecules containing 6 TetO sites, with position 6 being closest to the promoter (two biological replicates). The dashed line indicates the average occupancy.
F) Schematic of the Simple Competition model describing a direct competition of nucleosomes and TFs for binding to DNA.
G) Schematic of the Nucleosome Destabilization model describing a TF-dependent recruitment of a cofactor that reduces the binding energy of a nucleosome when one or more TFs are present.
H) Best fit parameters and error (SEM) for binding of rTetR-VP48 using Nucleosome Destabilization Model in (G).
I) Full molecular state representations for 10,000 measured (left) or simulated (according to the Nucleosome Destabilization, right) 6xTetO enhancer molecules, where each column represents a TetO site and each row represents a molecule. Sites are colored by their occupancy status.
J-K) Observed average occupancy (J) and occupancy distribution (K) of rTetR (only) (two biological replicates) with a fit from the Simple Competition model (r2 = 0.77 and 0.94, respectively). Gray line is rTetR-VP48 data for comparison. L-M) Observed average occupancy (L) and occupancy distribution (M) of rTetR-VP48 in the presence of the BAF inhibitor BRM014 (two biological replicates) with a fit from the Nucleosome Destabilization model (r2 = 0.94 and 0.96, respectively). Gray line is rTetR-VP48 data for comparison.
N-O) Observed average occupancy (N) and occupancy distribution (O) of rTetR-VP48 in the presence of the p300 inhibitor A485 (two biological replicates) with a fit from the Nucleosome Destabilization model (r2 = 0.99 and 0.90, respectively). Gray line is rTetR-VP48 data without inhibitors for comparison.
P) Parameter fits for models in L-O. Error bars represent the standard error of the mean from two biological replicates. Gray dashed line is rTetR-VP48 parameter fits for comparison.