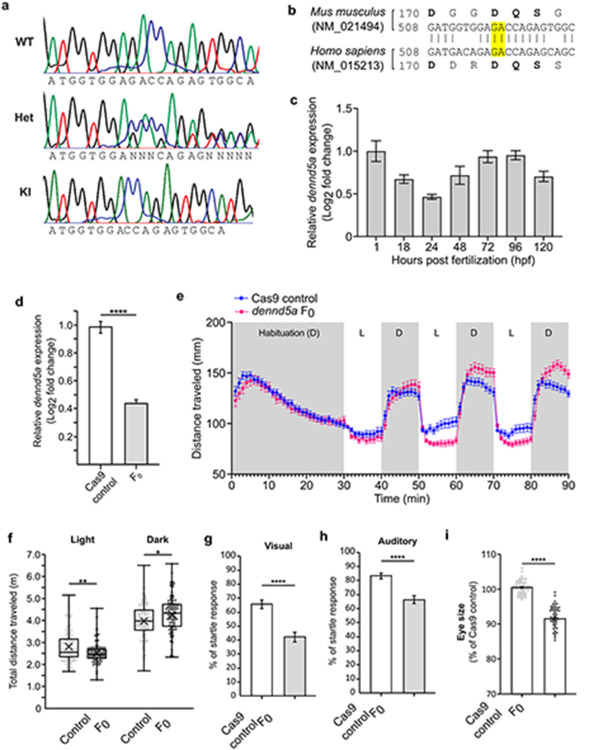
Extended Data Figure 5: Features of DENND5A transgenic animals make them valid models to study DENND5A-related DEE.
a, Sample chromatograms demonstrating DENND5A DNA sequences in WT, heterozygous (Het), and knock-in (KI) mice. b, DNA and amino acid sequence alignment between human and mouse DENND5A sequences. Highlighted base pairs indicate bases deleted using CRISPR/Cas9. c, The temporal expression of zebrafish dennd5a mRNA by RT-qPCR at different developmental stages from experiments performed with biological and technical triplicates. Expression levels were normalized to the 18S housekeeping gene and compared to 1 hpf embryos. Error bars = mean ± SD. d, Expression of dennd5a mRNA in Cas9 controls and dennd5a F0 knockouts detected by RT-qPCR at 5 dpf. Experiments were performed with four biological replicates with technical triplicates. Data are mean ± SEM analyzed via two-tailed student’s t-test (t(6) = 10.706, p < .0001). e, Locomotor activities of zebrafish larvae at 5 dpf with n = 96 larvae for each group. Data are mean ± SEM. D = Dark period, L = light period. f, Quantification of distance traveled by each larva during the cycles of light or dark periods analyzed via two-tailed Mann-Whitney U test (light; Z = −2.81, p = .005) and two-tailed student’s t-test (dark; t(190) = −2.438, p = .016). Each dot represents one larva. g, Visual startle response in n = 143 larvae at 6 dpf. Data are mean ± SEM analyzed via two-tailed Mann-Whitney U test (Z = −4.957, p < .0001). h, Acoustic evoked behavioral response in n = 134 larvae at 6 dpf. Data are mean ± SEM analyzed via two-tailed Mann-Whitney U test (Z = −4.947, p < .0001). i, Quantification of eye size in n = 60 larvae. Each dot represents one larva. Data are mean ± SEM analyzed via two-tailed Welch’s t-test (t(96.016) = 17.831, p < .0001).