Summary
Early detection is key for increased survival in ovarian cancer, but no general screening program exists today due to lack of biomarkers and overall cost versus benefit over traditional clinical methods. Here, we used dried cervico-vaginal fluid (CVF) as sampling matrix coupled with mass spectrometry for detection of protein biomarkers. We find that self-collected CVF on paper cards yields robust results and is suitable for high-throughput proteomics. Artificial intelligence–based methods were used to identify an 11-protein panel that separates cases from controls. In validation data, the panel achieved a sensitivity of 0.97 (95% CI 0.91–1.00) at a specificity of 0.67 (0.40–0.87). Analyses of samples collected prior to development of symptoms indicate that the panel is informative also of future risk of disease. Dried CVF is used in cervical cancer screening, and our results opens the possibility for a screening program also for ovarian cancer, based on self-collected CVF samples.
Subject areas: Women’s health, Body substance sample, Fluid, Cancer, Proteomics
Graphical abstract
Highlights
-
•
Dried cervico-vaginal fluid (CVF) on paper cards is suitable for proteomics
-
•
The CVF proteome is heterogeneous, based on mass spectrometry data
-
•
Several biomarker candidates for ovarian cancer were found in the CVF proteome
Women’s health; Body substance sample; Fluid; Cancer; Proteomics
Introduction
Ovarian cancer has an estimated global incidence of 6.6 per 100,000 women per year making it the 8th most common cancer among women across the world with over 300,000 cases reported and over 200,000 deaths.1 Detection of the cancer is usually symptom-driven, resulting in that less than one-third of cases are discovered early, in stage I or II, leading to a poor prognosis with an overall 5-year survival rate of only 30–50%.2 1Early detection of ovarian cancer could be facilitated by knowledge of the etiology of the cancer both by discovering precise biomarkers and determining an optimized screening interval in relation to cancer development. However, the precursor states of ovarian cancers have proven difficult to identify. Recent investigations have suggested that serous tubal intraepithelial carcinomas (STIC), a presumed precursor to high-grade serous ovarian carcinomas, develop slowly over up to two decades from the first occurrence of genetic predisposing mutations.3 Recent molecular evidence, however, from patient material suggests that ovarian cancer can develop from STIC in a much shorter time, as rapidly as in an estimated time span of 6–7 years.4,5 Other estimates based on tumor sizes and growth6 indicate that ovarian cancer can spend over 4 years in situ, or as stage I and II, before progressing to stages III and IV. The ovarian cancer diagnose is typically symptom-driven and women who experience pelvic symptoms are usually first examined with computed tomography or transvaginal ultrasound (TVU), and when these indicate an adnexal ovarian mass, surgery is used for a definitive diagnose. However, the majority of symptomatic patients undergoing surgery do not have malign tumors but normal or benign cysts7 and more effective and targeted preoperative tools to predict malignancy could reduce unnecessary surgery and minimize morbidity and induced premature menopause. A recent study analyzing copy number variation at single cell level in histologically defined benign and malign tissue across multiple cancers such as prostate cancer, ductal breast cancer and squamous cell carcinomas suggest that, at least for the studied cancers, copy number alterations present in benign tissue could precede tumorigenesis.8 Therefore, from a screening perspective, in order to identify all cancers at the earliest possible state, it is important to include also women that are examined due to suspicion of ovarian cancer but diagnosed with benign conditions.
Today, to the best of our knowledge, no molecular test accurate enough to justify population-wide screening of ovarian cancer has been described. The largest running ovarian cancer screening study, the United Kingdom Collaborative Trial of Ovarian Cancer Screening (UKCTOCS)9 uses multi-modal screening where indications of elevated MUCIN-16 (Cancer antigen-125/CA125) triggers a follow-up with TVU. A recent evaluation of the long-term results from the study10 concluded that although an increase could be observed in early-stage cancer detection, no clear reduction in mortality was detected among women participating in screening as compared to the un-screened group. Another large study with a similar screening strategy, the Normal Risk Ovarian Screening Study (NROSS), could however draw more promising conclusions with a larger shift toward earlier discovery.11 One remaining issue with the multi-modal approach however is the lower sensitivity of the biomarkers used as first line indicator, while a high specificity is obtained with TVU.11 Therefore, additional biomarkers with a higher sensitivity for early-stage ovarian cancer are needed. Few clinical biomarkers for ovarian cancer are however available today. MUCIN-16 was introduced as a biomarker for ovarian cancer in 198312 and is in most countries currently the only biomarker for diagnosis and management of ovarian cancer.13 However, MUCIN-16 alone has low sensitivity for early-stage cancer partly due to a large proportion of false positives resulting from non-cancer gynecological conditions such as endometriosis, infections, or normal pregnancies.13 Risk-scores using combinations of MUCIN-16 and other biomarkers such as the WAP four-disulfide core domain 2 (WFDC2 or HE4) used in the ROMA Score (Ovarian Malignancy Risk Algorithm), can achieve sensitivities of 90–95%14,15 at a specificity of 75%. A lower sensitivity for detection of early-stage ovarian cancer (stages I and II), high costs and risk of over-treatment due to low specificity prohibits population screening using these biomarkers. We have previously shown16 that combinations of plasma protein biomarkers can separate benign tumors from ovarian cancers at a very high sensitivity and specificity and that different biomarkers were optimal for separating early or late stages from benign conditions.16 Interestingly, in that study, the best performing biomarker combinations did not include MUCIN-16 suggesting that broad studies taking advantage of high-throughput proteomics technologies without prior assumptions on what biomarkers to include could detect novel biomarkers that outperform current gold-standards.
Self-sampled cervico-vaginal fluid (CVF) deposited on the FTA elute filter paper cards have been shown to provide accurate and cost-efficient screening of cervical cancer.17,18 We have previously also shown that dried CVF on paper cards is suitable for high-throughput proteomics using both affinity-based methods19 and mass spectrometry.20 A recent study by Boylan et al.21 analyzed the proteomic spectra with mass spectrometry in tumor tissue and a cervical swab from a woman diagnosed with late stage (metastatic), high-grade serous adenocarcinoma of ovarian or peritoneal origin. They found a large overlap in the detection of proteins between the two sample types, with 84.6% (3716 of 4392) and 88.6% (3716 of 4194) of the proteins commonly detected in the tumor and cervical swab respectively. This indicates that CVF could reflect the expressed tumor proteome and therefore be a useful sampling matrix also for detection of ovarian cancer.
Here, we aimed to evaluate dried CVF on paper cards as sampling method for early detection of ovarian cancer. To accomplish this, we have analyzed samples of dried CVF deposited on paper cards with mass spectrometry from both women surgically diagnosed with benign, borderline and malign tumors, healthy controls and in samples collected up to four years before diagnose. Our results indicate that the CVF-proteome is highly heterogeneous between individuals, that MUCIN-16 specifically is not commonly detected but, that there are promising protein biomarker candidates for early detection of ovarian cancer.
Results
The CVF proteome is highly heterogeneous between individuals
A total of 160 dried CVF samples (Table 1; Figure S1, STAR Methods) deposited on the indicating FTA card collected from 155 women were included in the study. 104 of the samples were collected at time of diagnosis from women with a suspicion of ovarian cancer and surgically diagnosed with benign or malign conditions in the Sahlgrenska University Hospital in Göteborg, Sweden. The 104 samples were collected from 100 women. For two of the women three biological replicates were included to estimate intra-individual variation. An additional 16 samples were from 15 women participating in the cervical cancer screening in Uppsala, Sweden, that later were examined with suspicion of ovarian cancer. Among these samples, one woman had samples collected at separate time-points. Finally, 40 samples from symptom-free healthy women also participating in the same cervical cancer screening program, were used as controls in the present study. All samples were collected using a sampling brush (Viba brush) which was then applied to the colored area of the FTA elute micro card. The cards were then left to dry 5 min before closure and stored in room temperature. The mass spectrometry analyses were carried out in two phases, with the controls and a proportion of the cases in a first run (N = 60) and the remainder of the samples in a second run (N = 100). The data for the controls for the first run have previously been published.20 Both mass spectrometry runs were carried out at the same facility by the same staff (STAR Methods).
Table 1.
Participant characteristics
| Group | Diagnose | N | Agea | CA125 U/mlb |
|---|---|---|---|---|
| Controls | Healthy | 40 | 52.3 (3.8) | n.a. |
| OC-BDc | 16 | 48.7 (9.0) | n.a. | |
| Benign | 7 | 46.4 (10.2) | n.a. | |
| Borderline | 1 | 35 | n.a. | |
| - Stage I | 1 | 35 | n.a. | |
| Malign | 8 | 52.4 (6.0) | n.a. | |
| - Stage I | 2 | 49.8 (3.0) | n.a. | |
| - Stage II | 2 | 48.7 (11.5) | n.a. | |
| - Stage III | 3e | 54.4 (4.4) | n.a. | |
| - Stage IV | 1 | 57.8 | n.a. | |
| OC-ADd | 100 | 58.1 (14.4) | 95.0 (124.5) | |
| Benign | 38 | 54.3 (16.8) | 26.0 (17.8) | |
| Borderline | 10 | 51.0 (14.2) | 73.0 (63.0) | |
| - Stage I | 10 | 51.0 (14.2) | 73.0 (63.0) | |
| Malign | 52 | 62.3 (11.8) | 421.5 (494.4) | |
| - Stage I | 15 | 62.5 (13.2) | 93.0 (83.0) | |
| - Stage II | 5 | 67.0 (10.6) | 71.0 (57.8) | |
| - Stage III | 20 | 60.2 (13.7) | 889.0 (738.3) | |
| - Stage IV | 12 | 63.5 (6.7) | 774.0 (285.4) |
Reported as mean (std. dev) at sample collection.
Clinically measured MUCIN-16 (CA125) reported as median (median abs. dev.) at time of diagnosis.
OC-BD, samples with ovarian cancer suspicion collected before diagnosis.
OC-AD, samples with ovarian cancer suspicion collected at time of diagnosis.
Two samples from the same women collected at different time-points. n.a: not available.
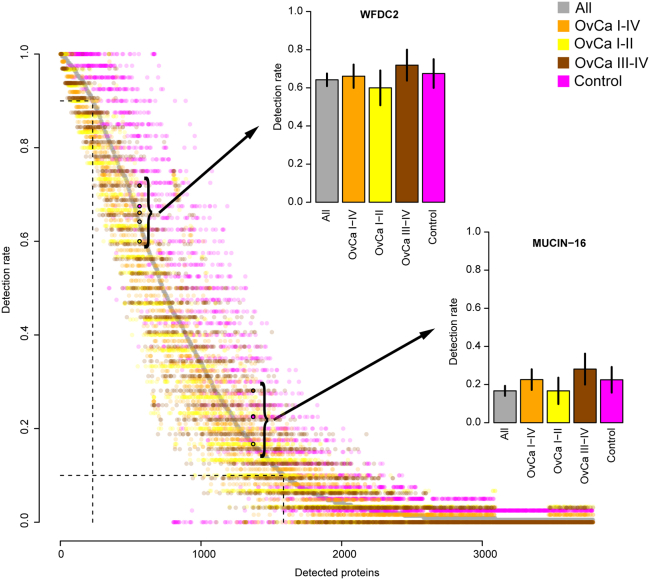
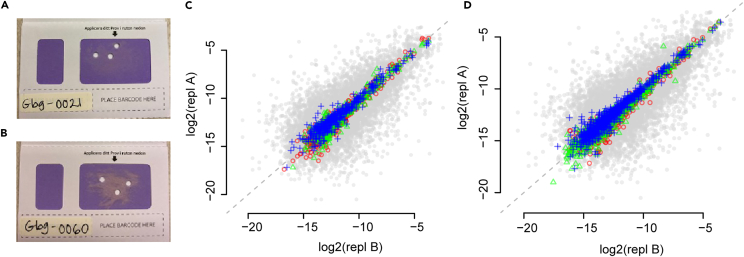
A total of 3788 proteins were detected in at least one sample (Figure 1; Table S1). The detection rate of proteins varied greatly between individuals with 42% of the proteins detected in at least 10% of the samples, but only 6% of proteins present in at least 90% of the samples. Specifically, WFDC2 and MUCIN-16, were detected in 64% and 16% of all samples and in 58% and 18% of the ovarian cancer samples, respectively (Figure 1). A subset (N = 19) of the samples analyzed here have previously also been characterized22 with a complementary proteomics technology, the proximity extension assay (PEA) in both separated plasma as well as the CVF deposited on FTA-cards. The affinity-based PEA technology is highly sensitive and here, both MUCIN-16 and WFDC2 were detected in 100% of the CVF-samples. These data were used to analyze the correlation between these two proteins as measured by PEA in plasma and CVF. The correlation coefficients were 0.2 (p value 0.39) and 0.04 (p value 0.87) for WFDC2 and MUCIN-16 respectively, and even though a more sensitive assay can detect MUCIN-16 and WFDC2 to a higher degree in CVF, the low correlation likely prohibits similar use in ovarian cancer for these two biomarkers in CVF as compared to plasma. We next analyzed if the observed high heterogeneity could be attributed to different sampling location for the punches on the FTA-card. This intra-individual variance was examined by comparing results from multiple punches from the same card for two separate samples. Figures 2A and 2B shows two FTA-cards with deposited CVF including three 3.5mm punches. The estimated correlation coefficients between any two pairs of punches in the two sets of replicates were all above 0.92 (Figures 2C and 2D, Spearman’s Rho, p value < machine precision, 2.2 x 10−16) indicating that robust results are obtained regardless of where on the card the punches are taken. A comparison with randomly selected pairs of cards, repeated 50 times (Figures 2C and 2D, gray dots) obtained a mean correlation coefficient of 0.72 (+/− 0.12). Finally, we also analyzed if there was any general difference in protein specific detection rates obtained in the samples collected at time of diagnose compared to those collected before diagnose and found no difference (Figure S1).Based on these analyses, we concluded that the overserved heterogeneity in detection rates between individuals are most likely due to real differences in concentrations in combination with limits of detection in the technology user. The raw data were then normalized (STAR Methods) and quality-controlled for detection frequency on both protein and individual (women) level (STAR Methods). For the women with multiple biological replicates, only one (selected at random) replicate was included in the downstream analysis. The initial quality control consisted of two steps, first only proteins detected in at least 85% of the individuals were kept and then, only individuals that had measurements of at least 85% of those proteins were included in the proceeding analysis. After quality-control, 275 proteins in 139 samples were used for further analysis (Figure S1).
Figure 1.
Protein detection rate in CVF
Protein detection rate (y axis) for each detected protein (x axis, ordered by overall detection rate) for all samples in the study (gray), ovarian cancer samples (orange), ovarian cancers stage I and II at time of diagnosis (yellow), ovarian cancers (OvCa) stage III and IV at time of diagnosis (dark red) and controls (magenta). Proteins are ordered by the detection rate among all samples. The detection frequencies of WFDC2 and MUCIN-16 are shown both at circles with black borders and as additional panels. In these, horizontal lines on each bar represent the standard error of the mean (SEM). In the main panel, the dashed gray horizontal and vertical lines correspond to 90% and 10% detection rates among all the samples. Note that the “All” category includes additional categories (samples collected before diagnosis (benign and malign) and borderline ovarian cancers) that are not displayed as individual categories.
Figure 2.
Technical evaluation of dried CVF and comparisons with wet plasma
(A) and (B) Photo of FTA-collection card analyzed with deposited CVF including the 3 punches used in the analysis in (C) and (D).
(C) Pairwise comparison of 3 replicated punches (A) from different places of dried CVF on a paper card (colored) and pairwise comparison of randomly selected samples (gray). In addition to color, each pairwise comparison is plotted with a different symbol. The random selection was done by selecting a pair of samples from the same diagnose group as the replicated analysis. The distribution of random comparisons is based on 50 randomly selected pairs.
(D) as (C) but for the card shown in (B). In all analyses of replicated punches, the correlation-coefficients (Spearman’s Rho R) between any two pairs of punches are above 0.92.
Univariate biomarkers in CVF for early detection of ovarian cancer
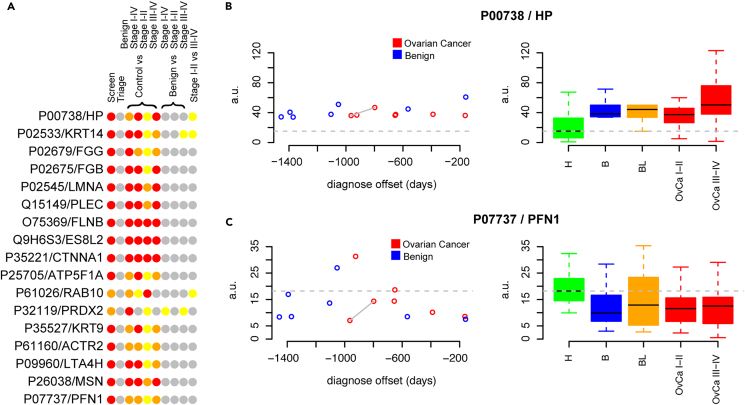
All 275 proteins passing quality control were analyzed univariately in a set of different comparisons depending on intended context. Firstly, we examined a screening scenario aiming to separate benign, borderline and malign tumors from controls, i.e., targeting the women that are today most commonly examined with suspicion of ovarian cancer. Secondly, we examined a triage scenario separating the benign tumors from borderline and malign tumors. We also examined the ability to separate; controls from benign, controls or benign from ovarian cancers (all stages), early-stage ovarian cancer (stage I and II) and late-stage cancers (stage III and IV). Lastly, we examined the ability to distinguish early-stage cancer form late-stage cancers. In total, we conducted 10 different comparisons for each protein and using a strict adjustment for multiple-hypothesis testing we found 17 proteins to be significantly different (p < 0.05/275/10 = 1.8 x 10−5) in at least one comparison with p values as low as 1.3 x 10−12 (Table S2; Figure 3A). The majority (15) of these 17 proteins had the lowest p value in the screening scenario with Peroxiredoxin-2 (UniProt P32119, controls vs. benign) and Ras-related protein Rab-10 (UniProt P61026, controls vs. early-stage ovarian cancer) being the two exceptions. Several of these 17 proteins however, displayed statistically different levels in multiple categories (Figure 3A). The proteins with the largest increase and decrease in mean proteins concentrations among the 17 detected biomarkers are shown in Figures 3B and 3C, respectively. Haptoglobin (Figure 3B right panel, UniProt P00738) shows an increase in benign, borderline and malign tumors compared to controls while Profilin-1 (Figure 3C right panel, UniProt P07737) shows a decrease. Focusing on the samples collected before diagnosis (Figure 3BC, left panel) a similar pattern was observed in terms of increased or decreased concentrations in relation to a future benign or malign diagnose. Haptoglobin (Figure 3B left panel) showed an increase compared to the controls in all samples (14/14) across the full-time span and as early as more than 1400 days prior to diagnosis. Similarly, 11 of 14 samples showed a decrease in cases as compared to controls for Profilin-1 across the full-time span (Figure 3C left panel).
Figure 3.
Univariate analysis of protein levels in CVF
(A) Schematic representation of nominally (yellow) significant, per-comparison multiple hypothesis adjusted (Bonferroni, orange) significant and overall multiple hypothesis adjusted (Bonferroni, red) significant different protein levels in the compared groups. Gray color represents non-significant associations (p > 0.05). Proteins are identified by UniProtID and ‘gene name’ as listed in the UniProt database.
(B) CVF-abundance levels of Haptoglobin (UniProt P00738) in samples collected before (left panel) and at time of diagnosis (right panel). The mean abundance level in controls (labeled ‘H’) is marked with a horizontal gray dashed line in both panels. In the right panel, ‘B’ represents women with benign conditions, ‘BL’ are borderline ovarian cancer, ‘OvCa I-II’ and ‘OvCa III-IV’ ovarian cancers stages I-II and III-IV respectively. The top and the bottom of the colored boxes represents the 25th and 75th percentile with the whiskers at 1.5x the interquartile range. The mean is indicated in each group with a solid black line. In the samples collected before diagnosis, the two data-points connected by a solid line are from the same woman at two different time-points.
(C) As (B) but for Profilin-1 (UniProt P07737).
A combination of 11 biomarkers in CVF for early detection of ovarian cancer
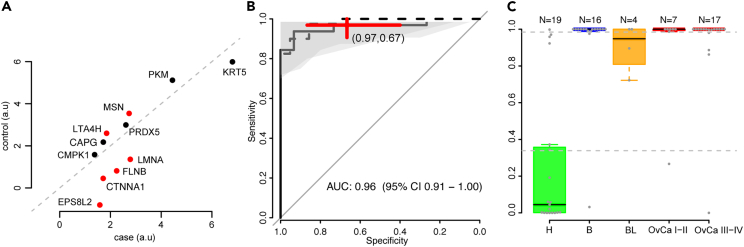
Next, we built a multivariate prediction model separating the controls from the benign, borderline and malign tumors at time of diagnosis. In this analysis, no missing values were allowed in the data why the data were first strictly filtered keeping 189 proteins in 100 samples. The filtering was done by removing either the individual or proteins with the highest missing value fraction until no missing values remained. Next, the data were split into a training (two-thirds, N = 62) and validation set (one-third, N = 41) keeping the fraction of cases and controls similar between the two sets. Only samples collected at time of diagnosis was used in the development of the multivariate model. The training fraction was then used to perform feature selection (STAR Methods) and to build a predictive model with a Naive Bayes classifier (STAR Methods) based on the proteins selected in the feature-selection step. The model returned a risk-score on the scale of 0–1 and a cut-off for malignancy was set representing a sensitivity of at least 95% in the training data. The feature-selection extracted 11 proteins (Figure 4A) out of which 6 were also found to be significant in the univariate analysis (Figure 4A). Six or the 11 proteins had, on average, higher concentrations in controls compared to cases (Figure 4A). A re-filtering of the individuals excluded in the very first filtering step was then done but examining only the 11 selected proteins and samples with no missing values for these 11 proteins were included in the validation data, which brought the total sample count in the validation to 47. The model was then applied to the samples in the validation data calculating the risk-score and subsequently predicted as control or case using the previously defined cut-off. We found no statistical difference (DeLong’s method, p = 0.59) in the area under curve (AUC) estimates of the receiver operating characteristics (ROC) obtained during training of the model (AUC = 0.98, 95% confidence interval (CI) 0.95 to 1.00) compared to the validation proportion of the data (AUC = 0.96, 95% CI 0.91–1.00, Figure 4B). In addition, there was no statistical difference in the sensitivity (p = 0.66, Fisher’s exact test) and specificity (p = 0.24, Fisher’s exact test) obtained in the validation proportion of the data at the specific cut-off developed in the training data. In the validation proportion of the data, the point-estimate of sensitivity and specificity of the model and cut-off were 97% and 67% respectively. The distribution of the risk-score in the validation proportion of the data can be seen in Figure 4C. We also calculated the risk-score for the samples collected prior to diagnosis with complete observations for the 11 proteins (12 samples) and could conclude that all samples were above the cut-off across the full time span with no significant difference between the obtained risk-scores (p = 0.81, two-sided Wilcoxon ranked based test) in the group that later was diagnosed with ovarian cancer compared to benign conditions. Finally, we analyzed if any of the 11 proteins in the multivariate model were influenced by individual age. Using all samples, we found no such influence (all p > 5.7 x 10−2, Table S3). When analyzing the three major groups (controls, benign and malign) separately we found 3 of the proteins to have nominally significant influence by age in the benign and control groups out of which one remained significant after adjustment for multiple hypotheses testing. The remaining significance was detected for pyruvate kinase PKM (UniProt ID P14618) in controls (p = 1.8 x 10−4) with higher levels observed with increasing age. There was a nominally significant association in benign samples (p = 0.03) while no association was detected in the malign group (p = 0.25).
Figure 4.
Multivariate risk-score for screening
(A) Mean protein abundance levels in the healthy controls (y axis) and the benign, borderline and malign tumors (x axis) for the 11 proteins in the risk-score. Red colored dots represent univariate significance with multiple hypothesis adjusted p values below 1.8 x 10−5.
(B) Receiver operating characteristics (ROC) curve for the predictions based on the risk-score at time of diagnosis. The dashed line corresponds to estimated performance during training (cross-validation) and the solid line the performance achieved in the validation proportion of the data using the trained model. The shaded gray areas correspond to 95% confidence interval in the validation proportion. The center of the red cross corresponds to the point-estimated sensitivity and specificity in the validation proportion of the data using the model and cut-off used in the training data. The width and height of the cross illustrate the 95% confidence interval of the estimated sensitivity and specificity.
(C) Distribution of the risk-score in specific groups of the validation data; H (healthy controls), B (benign), BL (borderline), OvCa I-II (ovarian cancer stage I and II) and OvCa III-IV (ovarian cancer stage III and IV). The number of samples in each group is specified at the top. Each individual sample is drawn as a solid gray dot with the top and the bottom of the colored boxes representing the 25th and 75th percentile and the whiskers at 1.5x the interquartile range. The median is indicated in each group with a solid black line. The horizontal dashed line corresponds to the cut-off established using the training data.
Discussion
Here, we have evaluated dried CVF on paper cards for use in early detection of ovarian cancer. Our results show that although the CVF-proteome as characterized by mass spectrometry is highly heterogeneous, several promising protein biomarkers for early detection were found. Based on proteins robustly detected across individuals, we found a combination of 11 proteins that demonstrated accurate, reproducibly, classification of cases and controls. An analysis of sample collected before diagnose indicate that this signal could also be detected prior to symptom driven discovery. New and accurate biomarkers for early discovery of ovarian cancer are urgently needed to increase survival.11 Biomarkers with low sensitivity leads to cancers being missed while a low specificity results in false positives, and possible over-treatment such as unnecessary surgery affecting fertility or introducing premature menopause. A functional screening program also needs to have a clear impact on the treatment of the disease and should lead to increased survival and quality of life for the affected. For ovarian cancer, there is a clear increase in survival when the cancer is detected early as compared to in later stages. Previous studies indicate that it could be possible to detect the cancer early, as the cancer pre-stages exists several years, or even decades, before developing into cancer.3,4,5,6 Given the relatively low population incidence for ovarian cancer, building prospective cohorts with samples collected prior to diagnosis is costly and requires long-time commitment of the participants to visit health care centers for donation of blood samples. Blood samples commonly used for biomarker studies, such as separated plasma or sera, also need dedicated biobanks for storage, handling and extraction requiring trained personnel, dedicated facilities, and long-term financing.23,24 Sampling by liquid biopsy, especially as self-collected samples, has the potential to allow participants to collect the sample when it suits them best, and to reduce cost for the health-care system.18 Collecting CVFs such as used here, is a minimally invasive sampling method that can be self-administrated. Another method that is suitable for self-administrated sample collection is dried blood samples (DBS). We have shown that in DBS samples from new-born screening stored at −24°C for up to 30 years, 75% of analyzed proteins remain stable over time.25 We have also reported that 17% of proteins analyzed in plasma display significant changes in abundance levels when stored at −80°C as plasma for up to 30 years with a similar effect size as the individual age of the patient.26 This implies that cheap and efficient long-time dry storage of self-collected samples as sampling matrix is not only a feasible alternative to current state-of-the art biobanking of liquid clinical samples at −80°C but may have significant advantages not only in terms sample preparation, storage cost but also for stability of analytes.
However, protein biomarker analyses using DBS27 or dried CVF as presented here does show differences when compared to analyses conducted in coupled plasma and/or serum from the same individuals. Therefore, the collected sample types should be considered as representing different tissues when searching for biomarkers for early discovery of ovarian cancer. We20 and others28 have shown that the individual CVF-proteome as characterized by mass spectrometry is highly heterogeneous as compared to what can be expected in plasma. In a previous study carried out in 2014 by Boylan et al.,28 five residual Papanicolaou (Pap)-test fluid samples from healthy women were analyzed using mass spectrometry and a normal core CVF-proteome consisting of 153 proteins was defined based on presence in 4 out of 5 samples. In our analysis of 40 controls, 113 of these 153 proteins were detected in at least 80% of the samples. When selecting 5 of the 40 samples at random and requiring at least 80% detection rate, an average of 640 proteins were found across 1000 such selections (Figure S2) highlighting the very heterogeneous nature of the CVF-proteome and the difficulties in defining the normal state. Apart from biological variation, a possible explanation to the varied results between the two investigations could also be the sampling method, and sample type, where the 153 proteins were detected in cell-free residual pap test fluid collected in cervical cancer screening which is not the same sample type as used here. The pap test specifically aims to collect cells from the cervix compared to the collection here which aims to collect fluid from the vagina. In addition, the numbers obtained here are, however, not immediately comparable to the earlier 153 proteins, as the assaying technique has improved, and more proteins are routinely detectable. As an example, a recent study21 investigated in detail three different sample types (tumor tissue, Pap-smear fluid, and a cervical swab) from the same woman diagnosed with late-stage ovarian cancer and detected close to 5000 proteins across the three sample types. They also found that the majority of these proteins were commonly detected in all three types. Specifically, in that study, both MUCIN-16 and WFDC2 were detected in all three tissue types. In our study, neither MUCIN-16 nor WFDC2 was detected in CVF in more than 1 of 5 ovarian cancer patients (stage I-IV) using mass spectrometry. Although the highly sensitive PEA detected both proteins in 100% of the dried CVF-samples, we found no correlation between the plasma and the CVF levels from the same women at the same time point. This indicates that these two biomarkers, although robustly detectable in CVF with sensitive assays, are not directly usable as CVF biomarkers for ovarian cancer in the same way as in plasma.
Many of the biomarker candidates identified here in the univariate analysis have previously been implicated in ovarian cancer, but a fair comparison is difficult to accomplish since the majority of previous studies have been based on serum, plasma or peritoneal fluid29 rather than CVF. Haptoglobin for instance, had the largest increase in cases compared to controls in our study. Higher haptoglobin expression in hepatocellular carcinoma (HCC) tissue have previously been associated with improved survival rates in HCC30 and elevated levels of haptoglobin in plasma in relation to ovarian cancer has also been observed.31 On the other hand, a recent study by Garibay-Cerdenares et al.32 specifically investigated the functional role of haptoglobin in cancer development and concluded that the protein is expressed by ovarian cancer cells, but only after exposure to ascitic fluid. The two proteins with the strongest statistical support in our study, (EPS8-related protein 2 (EPS8L2) and Catenin Alpha 1 (CTNNA1), both with p < 1.71 x 10−12) showed an increased expression in cases compared to controls. CTNNA1 has been indicated as a favorable prognostic maker for renal cancer while an unfavorable prognostic marker for pancreatic cancer based on gene expression in each respective tissue.33 Reduced expression in tumor tissue of CTNNA1 has been suggested to be a marker for early-stage ovarian epithelial cancers with poor prognosis,34 while decreased protein expression in different ovarian cancer cell lines have been shown to either increase or decrease sensitivity to different chemotherapies.35 A recent review36 of the role of EPS8, including the EPS8L2 homolog, reports overexpression (as seen here) in a large variety of cancers, including ovarian cancer with involvement in tumorigenesis, tumor proliferation, migration, metastasis as well as drug resistance and poor prognosis. Although several candidate biomarkers were found with strong statistical support comparing cases and controls in the screening scenario, we found no strong univariate candidates separating the benign tumors from malignant. A total of 25 biomarkers (Table S2) were nominally statistically significant (p < 0.05) but non remained significant after adjustment for multiple hypothesis testing (p < 1.8 x 10−5). The protein with the strongest statistical support in this comparison, the Eukaryotic initiation factor 4A-I (EIF4A1) were found to have higher levels in patients with benign tumors compared to malign (p = 1.9 x 10−3, Table S2). EIF4A1 has been implicated in several cancers,37 including gynecological cancers such as cervical and endometrial, but most often with higher expression in tumor tissue compared to normal associated with poor outcome. In summary, although there is good evidence from previous studies that the biomarker candidates detected here have cancer relevance, the interpretation is limited by the cross-tissue comparison and further investigations are needed to fully understand the functional relationships.
A major strength of our study is the use of samples collected on two locations, encompassing both healthy controls and women diagnosed with benign conditions in comparison to both early and late-stage ovarian cancer as well as samples collected up to four years prior to date of diagnosis. The use of standardized protocols for all molecular analyses carried out facilitate reproducibility of the conducted research, although more optimized protocols or assays could have increased the detectability of proteins across the analyzed samples. The analysis of multiple punches from the same women also strengthens our results in terms of reproducibility, as the obtained protein spectra from biological replicates are highly similar within individuals. The large inter-individual variance most likely reflects underlying biology with the protein concentrations among primarily low abundant proteins falling under the detection limit when analyzed with mass spectrometry. This was also observed for the subset of proteins with available data from the highly sensitive protein extension assay (PEA) where a 100% detection rate was obtained from dried CVF compared to lower detection rates with mass spectrometry. Future studies employing more sensitive technologies, such as antibody-based, are likely to obtain larger proteomics datasets in terms of higher detection rate across individuals and, subsequently, a larger selection from which biomarker candidates for early detection of ovarian cancer can be identified. This development has been evident in analyses conducted in plasma. For instance, in our own first study22 of 593 proteins in plasma we identified a biomarker panel consisting of 11 proteins with a sensitivity and specificity at 0.77 and 0.69 for separating benign conditions from early-stage ovarian cancer. In a subsequent study16 of 1472 proteins, a biomarker panel with 7 proteins achieved near perfect separation of the benign conditions from early-stage ovarian cancer, notably without MUCIN-16. On the other hand, technologies such as mass spectrometry offers a hypothesis free search for new biomarker candidates, while antibody-based technologies rely on a preselected set of analytes. In the current study, 11 of the 17 univariate biomarker candidates identified are not currently available among the over 5300 proteins in the commercial libraries of the PEA.
In conclusion, self-collected dried CVF deposited on paper cards is suitable for large scale proteomics with mass spectrometry and robust results are obtained regardless of where on the surface with CVF material the punch is taken from. Our analyses identified several novel biomarker candidates that could be valuable as first in line test for discovery of ovarian cancer. The same sample type as used here has been routinely used for screening of human papilloma viruses (HPVs) in cervical cancer screening, and our results opens the possibility for a future screening program in ovarian cancer utilizing the same type of self-collected samples.
Limitations of the study
Our study is limited by difficulties in obtaining samples collected at different timepoints prior to diagnosis, which could have been used to pinpoint a window in time where the signals start to deviate between cases and controls. Although a limited analyses, we found one of the proteins in the multivariate model to be influenced by individual age. This association was found in controls only. In our univariate analysis, this protein had higher levels in controls (Table S2) as compared to both benign and malign meaning that young enough healthy individuals could have similar levels of this protein as the cases. Our material however lacks the ages-span to analyze this in detail and further investigations are needed. We were also limited in functional interpretation of the biomarker candidates, as CVF represent a less commonly investigated tissue type. It is clear from both our work and the previous literature, that the behavior can be different between protein biomarkers in CVF and plasma. The study is also limited by that only Swedish samples were analyzed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Cervico-vaginal fluid | Homo sapiens | N/A |
| Plasma samples | Homo sapiens | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| LC/MS grade water | Fisher Chemical | Cat#51140 |
| DL-Dithiothreitol (DTT) | Sigma-Aldrich | Cat#43815 |
| 2-Iodoacetamide (IAA) | Sigma-Aldrich | Cat#I6125 |
| Sequencing Grade Modified Trypsin | Promega | Cat#V5111 |
| Acetonitrile, LC-MS grade, 99.8% | Fisher Chemical | Cat#047138.M1 |
| Formic acid | Sigma-Aldrich | Cat#F0507 |
| Ammonium bicarbonate, 99% | Fisher Chemical | Cat#393210010 |
| Critical commercial assays | ||
| Proximity Extention Assay, custom version. | Olink Proteomics AB | www.olink.com |
| Deposited data | ||
| Deposited data | This paper | https://doi.org/10.17044/scilifelab.24241237 |
| Data published in another paper | Gutiérrez, A. L. et al.20 Identification of Candidate Protein Biomarkers for CIN2+ Lesions from Self-Sampled, Dried Cervico–Vaginal Fluid Using LC-MS/MS. Cancers 2021, Vol. 13, Page 259213, 2592 (2021). | https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD026064 |
| Software and algorithms | ||
| MaxQuant software, version 1.5.1.2 | https://www.biochem.mpg.de/6304115/maxquant | https://www.maxquant.org/ |
| R version 4.2.2 | R Core Team (2022) | https://www.R-project.org/ |
| Adobe Illustrator version 27.9 | Adobe Systems Software Ireland Limited | https://www.adobe.com/products/catalog.html |
| Other | ||
| Viba-Brush | Rover Medical Devices BV, Oss, The Netherlands | https://www.roversmedicaldevices.com/cell-sampling-devices/viba-brush/ |
| FTA Elute Micro Card | GE Healthcare, Cardiff, UK | art. no WB129308 |
| Harris Micropunch 3 mm | Whatman | art.no WB100038 |
| Protein LoBind tubes 1.5 mL | Eppendorf | https://www.sigmaaldrich.com/SE/en/product/sigma/ep0030108116 |
| Speedvac system ISS110 | ThermoFisher Scientific, Massachusetts, USA | ThermoFisher Scientific |
| Q-Exactive Plus mass spectrometer | Thermo Finnigan, Bremen, Germany | https://www.thermofisher.com/order/catalog/product/IQLAAEGAAPFALGMBDK |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stefan Enroth (stefan.enroth@igp.uu.se).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Data. The mass spectrometry data for the controls in this paper has previously been deposited in the EMBL/PRIDE database under accession number PXD026064. The mass spectrometry data for the remainder of the analysed samples have been deposited in the Science for Life Laboratories (SciLifeLab) Data Repositories with the following identifier https://doi.org/10.17044/scilifelab.24241237.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
The samples come from two separate collections. The first set (N=40) was the cervical cancer screening program carried out in Uppsala, Sweden where self-sampling was applied. Details on these studies have been published before in Gustavsson et al. 201817 and Gustavsson et al. 2019.38 The samples from the cervical cancer screening cohort were collected between 2013 and 2015. From this sample collection we used 40 controls with eligibility defined as having negative HPV-status as characterized by the HpVIR-test,39 no overlap with the regional cancer registry in Uppsala, Sweden at any time before May 2021 with no records of surgery to the fallopian tubes, i.e. sterilization and salpingectomy or hysterectomy. From the same collection but from a non-overlapping set of women we also included 16 samples from 15 women that were HPV-negative at the screening sample but later (until May 2021) have been examined with suspicion of ovarian cancer after the cervical cancer screening sample was collected. These samples were collected between 159 and 1455 days before the final diagnose was made. It was not recorded when, in relation to this date, the first symptoms were noted. The second set of samples (100 women, a total of 104 samples including 2 cards with 3 punches each) were collected by a specialist in gynaecology at a tertiary centre for gynaecologic cancer surgery, Sahlgrenska University Hospital (Göteborg, Sweden) at time for admission to the hospital for surgery for a suspicious pelvic mass or ovarian cyst. During the vaginal speculum examination, the sampling brush (Viba brush) was inserted and rotated in the top of the vagina, vaginal fornices, and portio but not in the cervical canal. The brush was then applied to the coloured area of the FTA elute micro card and rotated one circle, left to dry five minutes before closure and stored in room temperature. This sample collection mimics the procedure and equipment used in our previous studies and in the cervical screening program.17,38 These samples were collected in 2015 and 2016 at time for admission 2-10 days prior to surgical intervention and diagnose. Inclusion criteria for this cohort was admission for surgery due to suspicion of ovarian pathology. Exclusion criteria for this cohort was surgery to the fallopian tubes, i.e. sterilization and salpingectomy or hysterectomy. Tumour stage and serum MUCIN-16 (CA125) values were extracted from accompanying medical journal records where available. Women with suspicion of ovarian cancer from both cohorts were surgically diagnosed with a benign, borderline or malignant histology. All samples were from Swedish women with a mean age of 55.7 years. Stratified ages for the different groups are given in Table 1. No genotype information was available for the participants. No information of ancestry/ethnicity nor gender was available for the participants. Informed consent was obtained from all participants. The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Regional Ethics Committee in Uppsala, Sweden (Dnr 2012/099) and Göteborg (Dnr: 201-15).
Method details
Mass spectrometry
Mass-spectrometry measurements for the controls (N=40) and a subset of the cases in the second sample set (N=20) have been carried out previously by the Mass Spectrometry facility at the Biomedical Centre, Uppsala, Sweden. The data from the controls were previously published in Gutiérrez et al.20 Here, the remainder of the second sample set (N=84) and the 16 samples collected before diagnosis were analysed at the same facility as used previously. The sample processing and LC-MS/MS measurements were carried out here exactly as described in Gutiérrez et al.,20 except for an update to the version of the UniProt database used in the final annotation of the proteins. In brief, one 3.5 mm punch was extracted from each sample. The proteins were reduced, alkylated and on-filter digested by trypsin according to a standard operating procedure. The collected peptide filtrate was vacuum centrifuged to dryness using a speedvac system. Dried peptides were resolved in 60 μL of 0.1% FA and further diluted 5 times prior to nano- LC-MS/MS. The peptides were separated in reversed-phase on a C18-column with 90 min gradient and electrosprayed on-line to a Q Exactive Plus mass spectrometer (Thermo Finnigan). Tandem mass spectrometry was performed applying Higher-energy C-trap Dissociation (HCD). The acquired data (RAW files) were processed using MaxQuant software, version 1.5.1.2, and database searches were performed using the implemented Andromeda search engine.40 MS/MS spectra were correlated to a FASTA database containing proteins from Homo sapiens extracted from the UniProt database (release date: May 2021). A decoy search database, including common contaminants and a reverse database, was used to estimate the identification false discovery rate where a rate of 1% was accepted. The search parameters included: maximum 10 ppm and 0.6 Da error tolerances for the survey scan and MS/MS analysis, respectively; enzyme specificity was trypsin; maximum one missed cleavage site allowed; cysteine carbamidomethylation was set as static modification; and oxidation of methionine was set as variable modification. The search criteria for protein identification were set to at least two matching peptides of 95% confidence level per protein.
Quantification and statistical analysis
First, the data from the two mass-spectrometry runs were merged based on the curated UniProtIDs as listed in the March 2022 release of the database. A total of 3788 proteins were detected in at least one sample across all the data. The data was then normalized on an individual level by dividing the label-free quantification (LFQ) measure by the total sum of LFQs for that sample. A per-protein batch-normalization was carried out between the two runs based on the mean abundance level among the samples from the second cohort. The normalization was done by adjusting the individual levels of the first cohort so that the mean became the same as in the second. For 2024 of the detected protein, there was no overlapping non-missing observations in either group to perform this normalization and these proteins were removed from further analysis. For this group of proteins, the mean percentage of samples with missing values were above 97% in each run. The remaining proteins were then quality controlled by requiring at least 85% non-missing observations across all samples for each protein. This filtering kept 275 proteins. We then required each sample to have non-missing observations for at least 85% of these proteins. This filtering removed 17 samples with individual detection frequencies ranging from 8.5 to 82.3% among the 275 proteins. The removed samples were all cases; 1 sample collected before diagnosis (malign) and 16 at diagnose (6 benign, 2 borderline, 9 malign (1 stage I, 3 stage II, 2 stage III and 2 stage IV). The final dataset consisted of 275 proteins in 139 samples.
The univariate analyses were performed one protein at a time, comparing two groups of measurements. A two-sided Wilcoxon ranked test was used to determine statistical significance. Here, 10 different comparisons were performed for each protein; (1) controls vs benign, borderline and malign tumours, (2) benign tumours vs borderline and malign tumours, (3) controls vs benign, (4) controls vs ovarian cancers (all stages), (5) controls vs early-stage ovarian cancer (stage I and II), (6) controls vs late-stage cancers (stage III and IV), (7) benign vs ovarian cancers (all stages), (8) benign vs early-stage ovarian cancer, (9) benign vs late-stage cancers and (10) early-stage cancer vs late- stage cancers. Three significance levels were defined, p-values < 0.05 as nominal significance, p-values < 0.05/279 = 1.8 x 10-4 as per-test multiple hypothesis adjusted significance and p-values < 0.05/279/10 = 1.8 x 10-5 as multiple hypothesis adjusted significance.
The multivariate analysis was done considering only non-missing measurements. Starting from the 275 proteins in the 139 individuals, the non-missing data was extracted by repeatedly removing the protein or individual with the highest missing fraction until no missing values remained. After this filtering, 189 proteins remained in 100 individuals (31 controls, 23 benign, 6 borderline, 30 malign (stage I-IV; 10,1,12,7), and 10 samples collected before diagnosis (4 benign and 6 malign). From these, 62 control, benign and malign samples were randomly selected (corresponding to two thirds) as training cohort with the remaining samples as validation cohort. A feature selection was done on training cohort based on recursive feature selection as implemented by the ‘rfe’ function in the ‘caret’ R-package41 (version 6.0.91) using ‘nbFuncs’ as functions with method set to ‘repeatCV’ with 4 repeats. This feature selection returned 11 proteins with an accuracy of 0.87 at a kappa of 0.75. We then re-evaluated the discarded individuals from the above missing-data filtering step specifically evaluating only the 11 selected proteins, resulting in 22 additional non-missing samples that were added to the validation cohort. A Naïve Bayes model was then trained in the training proportion of the data using the ‘caret’ R-package employing a three-fold cross-validation schema optimising the Laplace correction (‘fL’ parameter) from 0 to 1 in steps of 0.1, with and without kernel and bandwidth adjustment (‘adjust’ parameter) from 1 to 4 in steps of 0.1. The final model was built with ‘fL’ = 0, with kernels and ‘adjust’ = 2.4. The model returned a score in the range 0 to 1. A threshold (cut-off) for separating the classes was determined by evaluating the receiver operating characteristics (ROC) on the training cohort at a minimum sensitivity of 0.95, the threshold was set to 0.3384. The performance in the validation cohort compared to performance in the training cohort was estimated using the area under curve (AUC)-measure and the sensitivity and specificity achieved at the pre-determined threshold. Statistical difference in the AUC was determined with the DeLong’s test and a two-sided Fisher’s Exact test on the 2x2 matrix with true/false negative/positives was used to evaluate differences in achieved sensitivity and specificity at the pre-determined threshold. The ROC-curves were plotted using the pROC R-package42 (version 1.18.0). All other plots were made using standard R-functions. All analysis were carried out in R43 (version 4.2.2).
Acknowledgments
We are grateful for the willingness of the participants to donate samples for the research conducted here. This study was founded by the Lena Wäpplings Stiftelse (SE), Erik, Karin och Gösta Selanders Stiftelse (SE), Swedish Collegium for Advanced Study, Sjöbergstiftelsen, the Swedish Research Council (SE 2022-00857, U.G.), the Swedish Cancer Foundation (SE 220604FE, UG 190008PJ, KSu CAN 2018/384) and the Swedish state under the agreement between the Swedish government and the county council, the ALF-agreement (KSu). The funders had no role in the study design nor the decision to publish the results.
Author contributions
All authors read and approved the final version of the manuscript. Conceptualization: K.St., U.G., K.Su., and S.E. Data curation: A.W., E.I., and S.E. Formal Analysis: S.E. Funding acquisition: K.Su., U.G., and S.E. Investigation: J.H.L., A.W., E.I., I.G., and J.B. Methodology: G.S., J.B., and S.E. Project administration: S.E. Resources: K.St., U.G., K.Su., and J.B. Software: J.B. and S.E. Supervision: J.B. and S.E. Validation, Visualization, Writing – original draft: S.E. All authors: Writing – review and editing. S.E. and A.W. had full access to and verified the underlying data.
Declaration of interests
The authors declare no competing interests.
Published: January 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109001.
Supplemental information
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/CAAC.21660. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Trabert B., DeSantis C.E., Miller K.D., Samimi G., Runowicz C.D., Gaudet M.M., Jemal A., Siegel R.L. Ovarian cancer statistics, 2018. CA A Cancer J. Clin. 2018;68:284–296. doi: 10.3322/CAAC.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu R.C., Wang P., Lin S.F., Zhang M., Song Q., Chu T., Wang B.G., Kurman R.J., Vang R., Kinzler K., et al. Genomic landscape and evolutionary trajectories of ovarian cancer precursor lesions. J. Pathol. 2019;248:41–50. doi: 10.1002/PATH.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shih I.M., Wang Y., Wang T.L. The Origin of Ovarian Cancer Species and Precancerous Landscape. Am. J. Pathol. 2021;191:26–39. doi: 10.1016/J.AJPATH.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labidi-Galy S.I., Papp E., Hallberg D., Niknafs N., Adleff V., Noe M., Bhattacharya R., Novak M., Jones S., Phallen J., et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 2017;8:1093. doi: 10.1038/s41467-017-00962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown P.O., Palmer C. The preclinical natural history of serous ovarian cancer: Defining the target for early detection. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lycke M., Kristjansdottir B., Sundfeldt K. A multicenter clinical trial validating the performance of HE4, CA125, risk of ovarian malignancy algorithm and risk of malignancy index. Gynecol. Oncol. 2018;151:159–165. doi: 10.1016/j.ygyno.2018.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Erickson A., He M., Berglund E., Marklund M., Mirzazadeh R., Schultz N., Kvastad L., Andersson A., Bergenstråhle L., Bergenstråhle J., et al. Spatially resolved clonal copy number alterations in benign and malignant tissue. Nature. 2022:360–367. doi: 10.1038/s41586-022-05023-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs I.J., Menon U., Ryan A., Gentry-Maharaj A., Burnell M., Kalsi J.K., Amso N.N., Apostolidou S., Benjamin E., Cruickshank D., et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet. 2016;387:945–956. doi: 10.1016/S0140-6736(15)01224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menon U., Gentry-Maharaj A., Burnell M., Singh N., Ryan A., Karpinskyj C., Carlino G., Taylor J., Massingham S.K., Raikou M., et al. Ovarian cancer population screening and mortality after long-term follow-up in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet. 2021;397:2182–2193. doi: 10.1016/S0140-6736(21)00731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bast R.C., Han C.Y., Lu Z., Lu K.H. Next steps in the early detection of ovarian cancer. Commun. Med. 2021;1:36. doi: 10.1038/s43856-021-00037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bast R.C., Klug T.L., St John E., Jenison E., Niloff J.M., Lazarus H., Berkowitz R.S., Leavitt T., Griffiths C.T., Parker L., et al. A Radioimmunoassay Using a Monoclonal Antibody to Monitor the Course of Epithelial Ovarian Cancer. N. Engl. J. Med. 1983;309:883–887. doi: 10.1056/nejm198310133091503. [DOI] [PubMed] [Google Scholar]
- 13.Sölétormos G., Duffy M.J., Othman Abu Hassan S., Verheijen R.H.M., Tholander B., Bast R.C., Gaarenstroom K.N., Sturgeon C.M., Bonfrer J.M., Petersen P.H., et al. International Journal of Gynecological Cancer. Lippincott Williams and Wilkins; 2016. Clinical Use of Cancer Biomarkers in Epithelial Ovarian Cancer: Updated Guidelines from the European Group on Tumor Markers; pp. 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsen M.A., Sandhu N., Høgdall C., Christensen I.J., Nedergaard L., Lundvall L., Engelholm S.A., Pedersen A.T., Hartwell D., Lydolph M., et al. Evaluation of HE4, CA125, risk of ovarian malignancy algorithm (ROMA) and risk of malignancy index (RMI) as diagnostic tools of epithelial ovarian cancer in patients with a pelvic mass. Gynecol. Oncol. 2012;127:379–383. doi: 10.1016/j.ygyno.2012.07.106. [DOI] [PubMed] [Google Scholar]
- 15.Lycke M., Ulfenborg B., Kristjansdottir B., Sundfeldt K. Increased Diagnostic Accuracy of Adnexal Tumors with A Combination of Established Algorithms and Biomarkers. J. Clin. Med. 2020;9:299. doi: 10.3390/JCM9020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gyllensten U., Hedlund-Lindberg J., Svensson J., Manninen J., Öst T., Ramsell J., Åslin M., Ivansson E., Lomnytska M., Lycke M., et al. Next Generation Plasma Proteomics Identifies High-Precision Biomarker Candidates for Ovarian Cancer. Cancers. 2022;14:1757. doi: 10.3390/cancers14071757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustavsson I., Aarnio R., Berggrund M., Hedlund-Lindberg J., Strand A.S., Sanner K., Wikström I., Enroth S., Olovsson M., Gyllensten U. Randomised study shows that repeated selfsampling and HPV test has more than twofold higher detection rate of women with CIN2+ histology than Pap smear cytology. Br. J. Cancer. 2018;118:896–904. doi: 10.1038/bjc.2017.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aarnio R., Östensson E., Olovsson M., Gustavsson I., Gyllensten U. Cost-effectiveness analysis of repeated self-sampling for HPV testing in primary cervical screening: A randomized study. BMC Cancer. 2020;20:645–649. doi: 10.1186/S12885-020-07085-9/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berggrund M., Ekman D., Gustavsson I., Sundfeldt K., Olovsson M., Enroth S., Gyllensten U. Protein Detection Using the Multiplexed Proximity Extension Assay (PEA) from Plasma and Vaginal Fluid Applied to the Indicating FTA Elute Micro Card™. J. Circ. Biomark. 2016;5:9. doi: 10.5772/64000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutiérrez A.L., Lindberg J.H., Shevchenko G., Gustavsson I., Bergquist J., Gyllensten U., Enroth S. Identification of Candidate Protein Biomarkers for CIN2+ Lesions from Self-Sampled, Dried Cervico–Vaginal Fluid Using LC-MS/MS. Cancers. 2021;13:2592. doi: 10.3390/CANCERS13112592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boylan K.L.M., Afiuni-Zadeh S., Geller M.A., Argenta P.A., Griffin T.J., Skubitz A.P.N. Evaluation of the potential of Pap test fluid and cervical swabs to serve as clinical diagnostic biospecimens for the detection of ovarian cancer by mass spectrometry-based proteomics. Clin. Proteonomics. 2021;18:4–12. doi: 10.1186/S12014-020-09309-3/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enroth S., Berggrund M., Lycke M., Broberg J., Lundberg M., Assarsson E., Olovsson M., Stålberg K., Sundfeldt K., Gyllensten U. High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer. Commun. Biol. 2019;2:221. doi: 10.1038/s42003-019-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doucet M., Yuille M., Georghiou L., Dagher G. Biobank sustainability: current status and future prospects. J. Biorepository Sci. Appl. Med. 2017;5:1–7. doi: 10.2147/BSAM.S100899. [DOI] [Google Scholar]
- 24.Albert M., Bartlett J., Johnston R.N., Schacter B., Watson P. Biobank bootstrapping: Is biobank sustainability possible through cost recovery? Biopreserv. Biobanking. 2014;12:374–380. doi: 10.1089/bio.2014.0051. [DOI] [PubMed] [Google Scholar]
- 25.Björkesten J., Enroth S., Shen Q., Wik L., Hougaard D.M., Cohen A.S., Sörensen L., Giedraitis V., Ingelsson M., Larsson A., et al. Stability of proteins in dried blood spot biobanks. Mol. Cell. Proteomics. 2017;16:1286–1296. doi: 10.1074/mcp.RA117.000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enroth S., Hallmans G., Grankvist K., Gyllensten U. Effects of Long-Term Storage Time and Original Sampling Month on Biobank Plasma Protein Concentrations. EBioMedicine. 2016;12:309–314. doi: 10.1016/j.ebiom.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broberg K., Svensson J., Grahn K., Assarsson E., Åberg M., Selander J., Enroth S. Evaluation of 92 cardiovascular proteins in dried blood spots collected under field-conditions: Off-the-shelf affinity-based multiplexed assays work well, allowing for simplified sample collection. Bioessays. 2021;43 doi: 10.1002/bies.202000299. e2000299–9. [DOI] [PubMed] [Google Scholar]
- 28.Boylan K.L., Afiuni-Zadeh S., Geller M.A., Hickey K., Griffin T.J., Pambuccian S.E., Skubitz A.P. A feasibility study to identify proteins in the residual Pap test fluid of women with normal cytology by mass spectrometry-based proteomics. Clin. Proteonomics. 2014;11:30. doi: 10.1186/1559-0275-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q., Zhang Z.H., Wang S., Lang J.H. Circulating cell-free DNA or circulating tumor dna in the management of ovarian and endometrial cancer. OncoTargets Ther. 2019;12:11517. doi: 10.2147/OTT.S227156. Preprint at Dove Medical Press Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tai C.S., Lin Y.R., Teng T.H., Lin P.Y., Tu S.J., Chou C.H., Huang Y.R., Huang W.C., Weng S.L., Huang H.D., et al. Haptoglobin expression correlates with tumor differentiation and five-year overall survival rate in hepatocellular carcinoma. PLoS One. 2017;12 doi: 10.1371/JOURNAL.PONE.0171269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shield-Artin K.L., Bailey M.J., Oliva K., Liovic A.K., Barker G., Dellios N.L., Reisman S., Ayhan M., Rice G.E. Identification of ovarian cancer-associated proteins in symptomatic women: A novel method for semi-quantitative plasma proteomics. Proteonomics Clin. Appl. 2012;6:170–181. doi: 10.1002/PRCA.201100008. [DOI] [PubMed] [Google Scholar]
- 32.Garibay-Cerdenares O.L., Hernández-Ramírez V.I., Osorio-Trujillo J.C., Gallardo-Rincón D., Talamás-Rohana P. Haptoglobin and CCR2 receptor expression in ovarian cancer cells that were exposed to ascitic fluid: exploring a new role of haptoglobin in the tumoral microenvironment. Cell Adhes. Migrat. 2015;9:394–405. doi: 10.1080/19336918.2015.1035504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Tissue-based map of the human proteome. Science. 2015;347:347. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 34.Anttila M., Kosma V.M., Ji H., Wei-Ling X., Puolakka J., Juhola M., Saarikoski S., Syrjänen K. Clinical significance of alpha-catenin, collagen IV, and Ki-67 expression in epithelial ovarian cancer. J. Clin. Oncol. 1998;16:2591–2600. doi: 10.1200/JCO.1998.16.8.2591. [DOI] [PubMed] [Google Scholar]
- 35.Miller H., Bassiouni R., Li Y., Roman L.D., Carpten J. Role of alpha catenin in ovarian cancer cell line sensitivity to platinum-based chemotherapy and PARP inhibitors. Gynecol. Oncol. 2020;159:94–95. doi: 10.1016/j.ygyno.2020.05.083. [DOI] [Google Scholar]
- 36.Luo K., Zhang L., Liao Y., Zhou H., Yang H., Luo M., Qing C. Effects and mechanisms of Eps8 on the biological behaviour of malignant tumours. Oncol. Rep. 2021;45:824–834. doi: 10.3892/OR.2021.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raza F., Waldron J.A., Quesne J.L. Translational dysregulation in cancer: eIF4A isoforms and sequence determinants of eIF4A dependence. Biochem. Soc. Trans. 2015;43:1227–1233. doi: 10.1042/BST20150163. [DOI] [PubMed] [Google Scholar]
- 38.Gustavsson I., Aarnio R., Berggrund M., Hedlund-Lindberg J., Sanner K., Wikström I., Enroth S., Olovsson M., Gyllensten U. Randomised study of HPV prevalence and detection of CIN2+ in vaginal self-sampling compared to cervical specimens collected by medical personnel. Int. J. Cancer. 2019;144:89–97. doi: 10.1002/ijc.31637. [DOI] [PubMed] [Google Scholar]
- 39.Gustavsson I., Aarnio R., Myrnäs M., Hedlund-Lindberg J., Taku O., Meiring T., Wikström I., Enroth S., Williamson A.L., Olovsson M., Gyllensten U. Clinical validation of the HPVIR high-risk HPV test on cervical samples according to the international guidelines for human papillomavirus DNA test requirements for cervical cancer screening. Virol. J. 2019;16:107. doi: 10.1186/s12985-019-1216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J. v, Mann M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn M. Building Predictive Models in R Using the caret Package. J. Stat. Software. 2008;28:1–26. doi: 10.18637/JSS.V028.I05. [DOI] [Google Scholar]
- 42.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77–78. doi: 10.1186/1471-2105-12-77/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Core Team . R Foundation for Statistical Computing; 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data. The mass spectrometry data for the controls in this paper has previously been deposited in the EMBL/PRIDE database under accession number PXD026064. The mass spectrometry data for the remainder of the analysed samples have been deposited in the Science for Life Laboratories (SciLifeLab) Data Repositories with the following identifier https://doi.org/10.17044/scilifelab.24241237.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.